|
产品名称 |
MOLM13人急性髓原白血病细胞 |
|
货号 |
ZQ0892 |
|
产品介绍 |
从一名20岁急性髓系白血病患者的外周血中建立,该患者在最初的骨髓增生异常综合征后于1995年复发(MDS,成纤维细胞过多的难治性贫血,RAEB);携带FLT3的内部串联复制;FLT3蛋白不表达;细胞系携带cell外显子8突变体;MOLM-14姊妹细胞系(ACC 777)。外显子组和核糖核酸序列数据可用(见参考文献18187和外胚层序列和RNA-Seq)。
MOLM13细胞具有悬浮生长特性,形态上接近淋巴样母细胞。该细胞系在体外培养时经过鉴定,能够保持稳定生长,且广泛应用于急性髓系白血病的基础研究和药物筛选中。特别是由于其携带FLT3突变,该细胞系常被用于FLT3靶向治疗的相关研究。MOLM13细胞系的外显子组和RNA测序数据已公开,可为AML相关的分子机制和治疗策略研究提供参考。MOLM13细胞系在急性髓系白血病病理研究和药物开发中占据重要地位。 |
|
种属 |
人 |
|
性别/年龄 |
男/20岁 |
|
组织 |
外周血 |
|
疾病 |
急性髓细胞白血病 |
|
细胞类型 |
肿瘤细胞 |
|
形态学 |
圆形 |
|
生长方式 |
悬浮 |
|
倍增时间 |
3-4 days (PubMed=9305600); 24 hours (PubMed=25984343); 41.1 hours (PubMed=28052028); ~50 hours |
|
培养基和添加剂 |
RPMI-1640(品牌:中乔新舟 货号:ZQ-200)+20%胎牛血清(中乔新舟 货号:ZQ0500)+1%P/S(中乔新舟 货号:CSP006) |
|
推荐完全培养基货号 |
|
|
生物安全等级 |
BSL-1 |
|
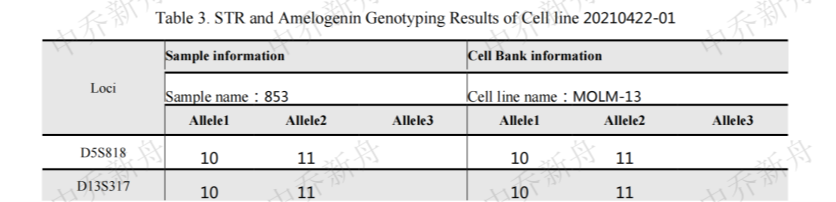
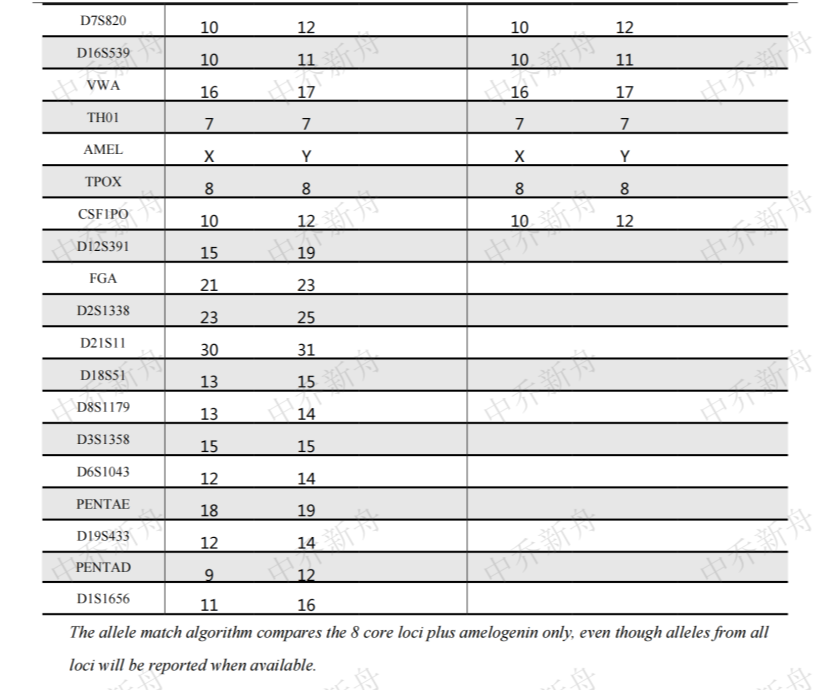
STR位点信息 |
Amelogenin X,Y CSF1PO 10 (AddexBio=C0003003/60) 10,12 (COG; Cosmic-CLP=1330947; DSMZ=ACC-554; JCRB=JCRB1810; PubMed=25877200) D2S1338 23,25 D3S1358 15 D5S818 10,11 D7S820 10,12 D8S1179 13,14 D13S317 10,11 D16S539 10,11 D18S51 13,15 D19S433 12,14 D21S11 30,31 FGA 21,23 Penta D 9,12 Penta E 18,19 TH01 7 TPOX 8 vWA 16,17 |
|
培养条件 |
95%空气,5%二氧化碳;37℃ |
|
抗原表达/受体表达 |
*** |
|
基因表达 |
*** |
|
保藏机构 |
DSMZ; ACC-554 JCRB; JCRB1810 |
|
供应限制 |
仅供科研使用 |
|
货号 |
ZQ0892 |
|
发货规格 |
活细胞:T25培养瓶*1瓶或者1ml 冻存管*2支(细胞量约为1x10^6 Cells/Vial)二选一 |
|
发货形式 |
活细胞:常温运输;冻存管:干冰运输 |
|
储存温度 |
活细胞:培养箱;冻存管:液氮罐 |
|
产地 |
中国 |
|
供应限制 |
仅供科研使用 |

 上海中乔新舟生物科技有限公司
上海中乔新舟生物科技有限公司