|
产品名称 |
SNU-449人肝癌细胞 |
|
货号 |
ZQ0787 |
|
产品介绍 |
SNU-449 于 1990 年由 J.-G. Park 及其同事在细胞毒治疗前取自一名韩国患者的原发性肝细胞癌。被广泛用于肝癌的研究,包括药物筛选、癌症生物学特性研究等。 |
|
种属 |
人 |
|
性别/年龄 |
男/52岁 |
|
组织 |
肝脏 |
|
疾病 |
癌; 肝细胞; II-III/IV 级 |
|
细胞类型 |
肿瘤细胞 |
|
形态学 |
上皮; 扩散的细胞 |
|
生长方式 |
贴壁 |
|
倍增时间 |
|
|
培养基和添加剂 |
RPMI-1640基础培养基(品牌:中乔新舟 货号:ZQ-200)+10%胎牛血清(中乔新舟 货号:ZQ500-A)+1%P/S(中乔新舟 货号:CSP006) |
|
推荐完全培养基货号 |
|
|
生物安全等级 |
BSL-1 |
|
培养条件 |
95%空气,5%二氧化碳;37℃ |
|
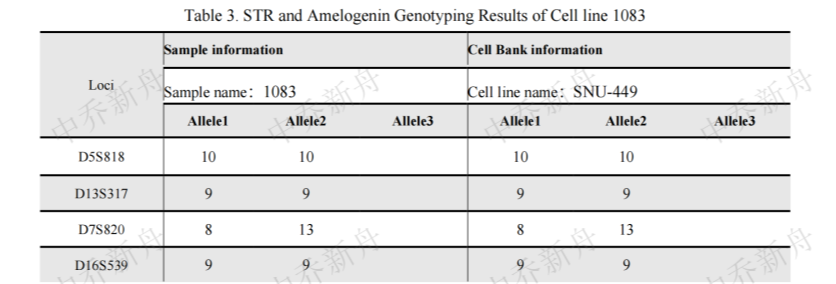
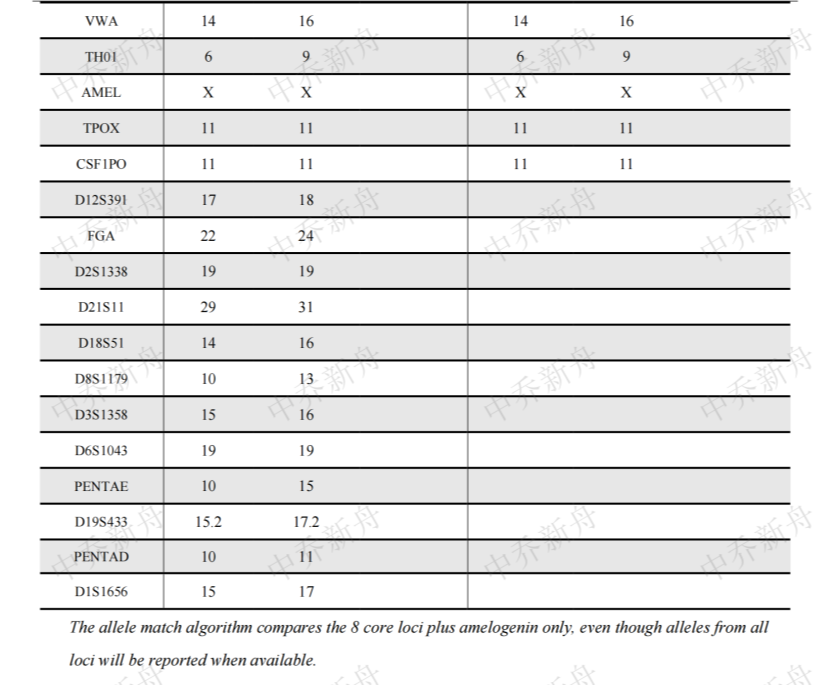
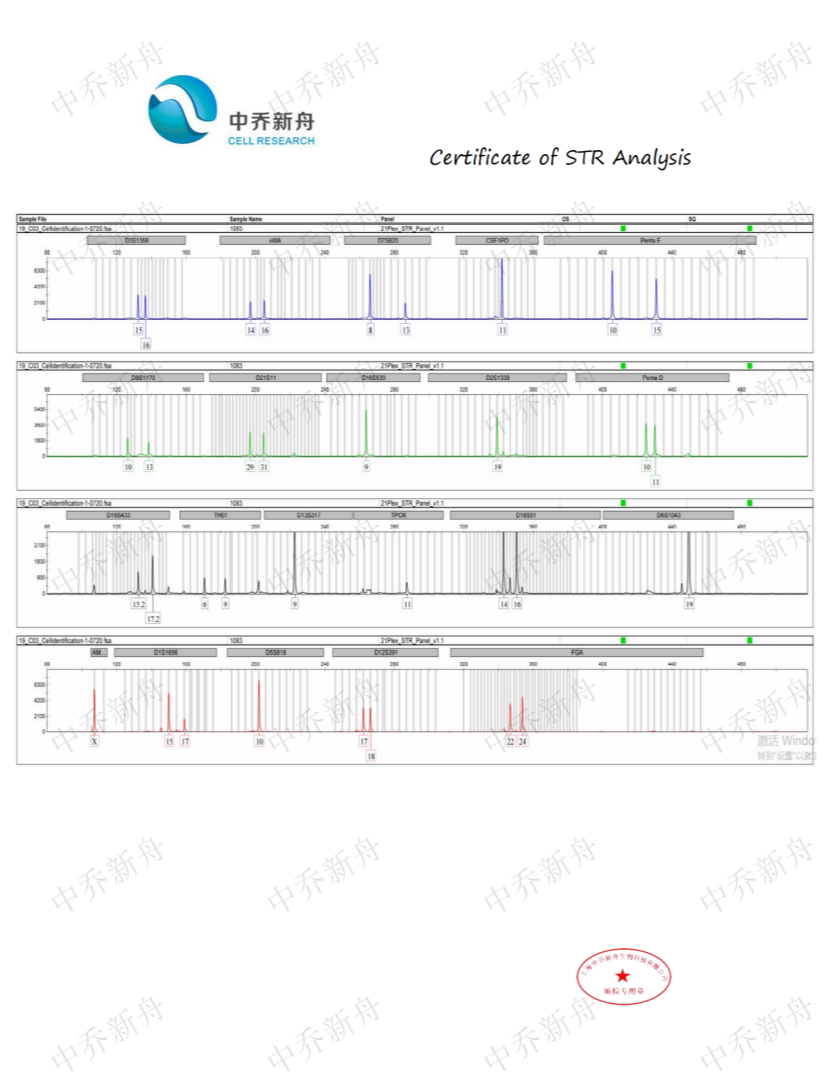
STR位点信息 |
Amelogenin: X,Y
CSF1PO: 11
|
|
抗原表达/受体表达 |
*** |
|
基因表达 |
*** |
|
保藏机构 |
ATCC; CRL-2234;KCLB; 00449 |
|
供应限制 |
仅供科研使用 |
|
货号 |
ZQ0787 |
|
发货规格 |
活细胞:T25培养瓶*1瓶或者1ml 冻存管*2支(细胞量约为1x10^6 Cells/Vial)二选一 |
|
发货形式 |
活细胞:常温运输;冻存管:干冰运输 |
|
储存温度 |
活细胞:培养箱;冻存管:液氮罐 |
|
产地 |
中国 |
|
供应限制 |
仅供科研使用 |
 上海中乔新舟生物科技有限公司
上海中乔新舟生物科技有限公司