|
产品名称 |
K-562人慢性髓原白血病细胞 |
|
货号 |
ZQ0424 |
|
产品介绍 |
K562细胞系来源于患有慢性骨髓性白血病的53岁女性患者的骨髓,Anderson等对其表面膜性质的研究表明,K-562是一株人类红白血病细胞。 K-562细胞株在自然杀伤分析中广泛作为高敏感的体外受体。 See Pross等将它用于NK细胞的体外详细检测,包括NK细胞活性的数学量化。 K-562母细胞是多能性,造血恶性细胞,它能自发分化成可识别的红血球祖细胞、粒性白细胞、单核细胞等。它是EBNA阴性的。是免疫学、肿瘤免疫学、免疫系统紊乱研究等多个领域的基础。人类K-562细胞广泛用于涉及免疫系统相互作用的研究,特别是与自然杀伤细胞(NK)等效应细胞的相互作用。这是由于它们的独特特性,例如表达可被NK细胞识别的特定抗原。探索NK细胞与K562等癌细胞系之间的相互作用,可以深入了解免疫防御机制。NK细胞对K562细胞的识别和反应能力随着特定标记物的存在而变化,这些标记物在整个K562细胞周期中波动。
注意事项: |
|
种属 |
人 |
|
性别/年龄 |
女/53岁 |
|
组织 |
骨;骨髓 |
|
疾病 |
慢性粒细胞白血病Cml |
|
细胞类型 |
肿瘤细胞 |
|
形态学 |
成淋巴细胞 |
|
生长方式 |
悬浮 |
|
倍增时间 |
大约17~47小时 |
|
常规培养基和添加剂 |
IMDM培养基(品牌:中乔新舟 货号:ZQ-900)+10%胎牛血清(中乔新舟 货号:ZQ0500)+1%P/S(中乔新舟 货号:CSP006) |
|
推荐含血清完全培养基货号 |
|
|
推荐无血清完全培养基货号 |
|
|
生物安全等级 |
BSL-1 |
|
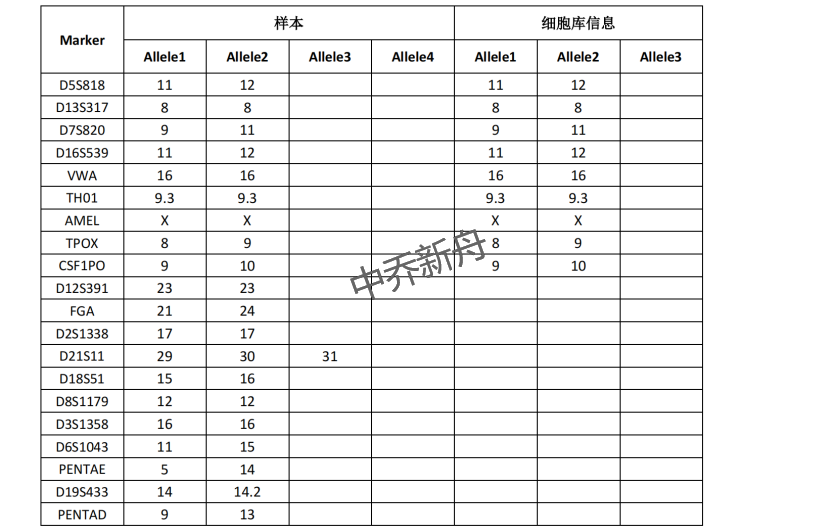
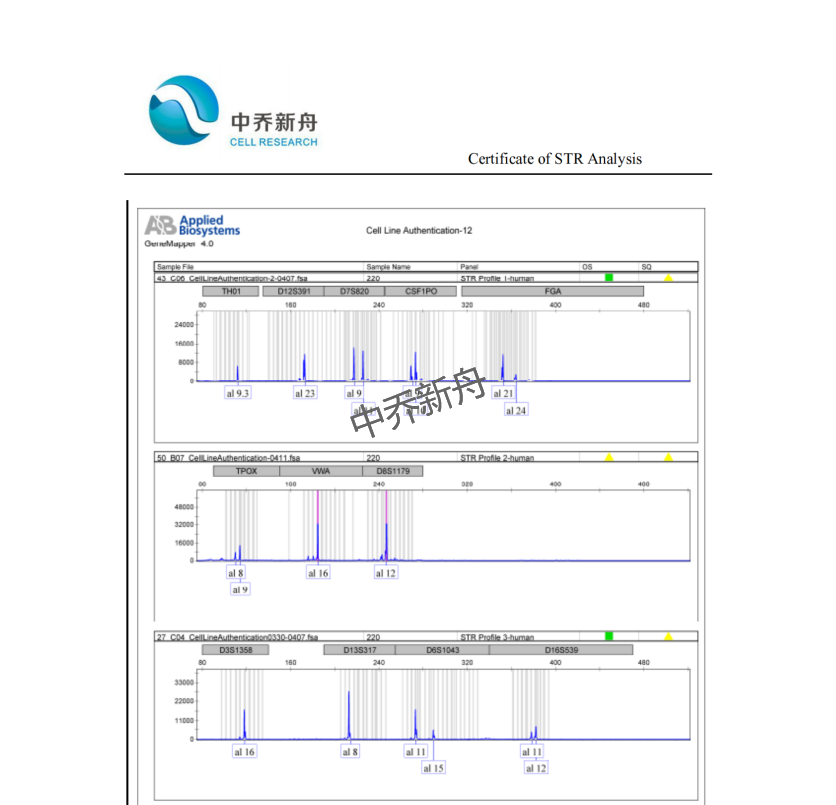
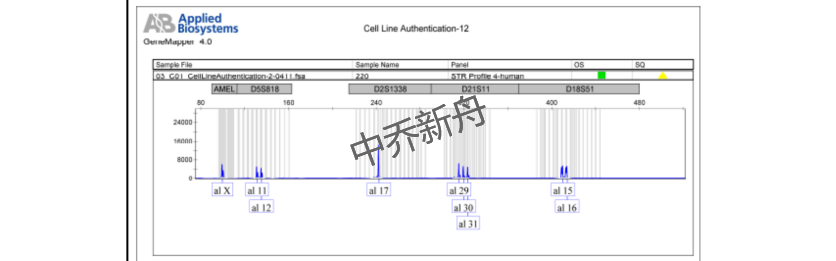
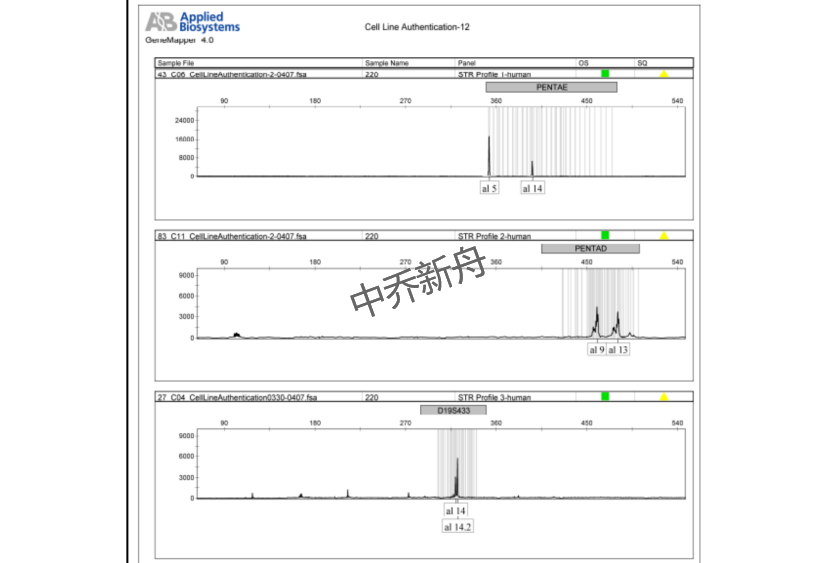
STR位点信息 |
Amelogenin: X CSF1PO: 9,10 D13S317: 8 D16S539: 11,12 D5S818: 11,12 D7S820: 9,11 THO1: 9.3 TPOX: 8,9 vWA: 16 |
|
培养条件 |
95%空气,5%二氧化碳;37℃ |
|
抗原表达/受体表达 |
*** |
|
基因表达 |
*** |
|
保藏机构 |
ATCC; CCL-243 BCRC; 60007 BCRJ; 0126 CCLV; CCLV-RIE 0439 DSMZ; ACC-10 ECACC; 89121407 |
|
供应限制 |
仅供科研使用 |
|
货号 |
ZQ0424 |
|
发货规格 |
活细胞:T25培养瓶*1瓶或者1ml 冻存管*2支(细胞量约为1x10^6 Cells/Vial)二选一 |
|
发货形式 |
活细胞:常温运输;冻存管:干冰运输 |
|
储存温度 |
活细胞:培养箱;冻存管:液氮罐 |
|
产地 |
中国 |
|
供应限制 |
仅供科研使用 |
| 货号 | 产品名称 | 规格 | 价格 | 指令 |
| ZQ505 | 支原体检测试剂盒(买二送一) | 1ml/100T | ¥800.00 | 放入购物车 》 |
| ZQ506 | 支原体清除试剂盒 | 1ml | ¥600.00 | 放入购物车 》 |
| A1001-1 | 国产转染试剂 | 1ml | ¥800.00 | 放入购物车 》 |
| 1 | 慢病毒介导基因沉默或过表达 | ¥询价 | 放入购物车 》 | |
| 6 | 绿/红色荧光蛋白标记 | ¥询价 | 放入购物车 》 | |
| ZQ500-A | 优级胎牛血清 | 500ml | ¥2580(开学促销) | 放入购物车 》 |
| 0513 | 双抗(青霉素/链霉素)溶液 | 100ml | ¥384.00 | 放入购物车 》 |
| 0563 | 支原体抑制剂 10 mg/ml; 1 ml(定制) | 10 mg/ml; 10 x 1 ml | ¥1808.00 | 放入购物车 》 |
| ZQ-900 | IMDM 基础培养基 | 500ml | ¥96.00 | 放入购物车 》 |
| ZQ-901 | IMDM 完全培养基(10%FBS) | 500ml | ¥350.00 | 放入购物车 》 |
| JSFW-134 | 人源细胞STR鉴定服务 | ¥900.00 | 放入购物车 》 |
 上海中乔新舟生物科技有限公司
上海中乔新舟生物科技有限公司