|
产品名称 |
MCF-7人乳腺癌细胞 |
|
货号 |
ZQ0071 |
|
产品介绍 |
MCF-7是一种人类乳腺癌细胞系,最初是从一位名叫Helen Marion的修女的胸腔积液中分离出来的。保留了乳腺上皮的分化特性,包括:能通过胞质雌激素受体加工雌二醇;含有Tx-4癌基因;肿瘤坏死因子α(TNFalpha)可以抑制MCF-7细胞的生长。抗雌激素处理细胞能调节IGFBPs的分泌。这个细胞系也因其在雌激素受体(ER)α研究中的重要性而被广泛使用,因为它是少数表达大量ER的细胞系之一,模仿了大多数侵袭性人类乳腺癌的特点。因其独特的生物学特性,在乳腺癌治疗研究中有着广泛的应用,例如:药物敏感性研究、基因组学研究、耐药性机制研究、基因功能研究、细胞信号传导研究、癌症转移研究、癌症干细胞研究、药物组合研究、精准医疗研究等。这些应用展示了MCF-7细胞系在乳腺癌治疗研究中的多样性和重要性,是乳腺癌研究的基石,它不仅帮助科学家们理解了乳腺癌的生物学特性,还促进了新治疗方法的发展。尽管存在局限性,但MCF-7细胞系预计仍将是未来乳腺癌研究的重要资源。 【注意事项】: 1. 该细胞贴壁较慢,建议复苏、传代后让细胞贴壁48-72h后再进行后续实验操作。 2. 使用0.25%(w/v) Trypsin-0.53 mM EDTA消化细胞。 3. MCF7是一种生长缓慢的细胞系,它会以松散的三维团簇的形式出现,并伴有一些漂浮的活细胞。悬浮的活细胞可以通过离心后重新加入到培养瓶中培养。在复苏后前二代该细胞会出现贴壁缓慢的情况,是正常现象。松散的三维团簇细胞慢慢会扩散形成一个扁平的单层细胞。 4. 如果生长速度比正常情况慢,可以尝试另一批FBS(即血清批次的促生长能力可能不同)。此外,将血清浓度增加到20%也可能有助于改善生长。 复苏时密度在8.0 x 104 - 1.5 x 105活细胞/cm2之间;传代时接种密度,在4.0 x 104 - 1.0 x 105活细胞/cm2之间。 一般来说,当开始培养MCF-7 时,如果观察到一些悬浮着的细胞或者小的细胞团,在传代时候,需要将这些悬浮的细胞或者小细胞团保留,跟贴壁细胞一起培养, 经过几天的培养后,这些细胞应该会重新贴附在培养瓶上并连接在一起,呈现叠层成岛状(dome)生长。随后,细胞会沿着这些岛边缘,向外生长扩散,在两次传代后,细胞应该会铺展开来,一般6-12天可以达到70-80%的密度。 对于T-25瓶培养, 客户首先按传代密度扩增,制备冻管5支左右,然后开始培养实验用细胞,当传代发现细胞增殖或形态异常时,终止传代,重新复苏细胞。实验用细胞控制传代频率,代数不宜超过5次。 |
|
种属 |
人 |
|
性别/年龄 |
女/69岁 |
|
组织 |
乳腺、胸;分离自转移灶:胸腔积液 |
|
疾病 |
非特殊类型的浸润性乳腺癌 |
|
细胞类型 |
肿瘤细胞 |
|
形态学 |
上皮细胞 |
|
生长方式 |
贴壁 |
|
倍增时间 |
大约24~80小时 |
|
培养基和添加剂(默认) |
MEM(含NEAA)(中乔新舟 货号:ZQ-300)+10%胎牛血清(中乔新舟 货号:ZQ0500)+1%双抗(中乔新舟 货号:CSP006)+1%L-alanyl-L-glutamine(中乔新舟 货号:CSP004)+1%SP(中乔新舟 货号:CSP003)+10ug/mL human recombinant insulin(中乔新舟 货号:CSP001-10) |
|
培养方案B(可选) |
DMEM (中乔新舟 货号:ZQ-100)+10%胎牛血清(中乔新舟 货号:ZQ500-A)+1%双抗(中乔新舟 货号:CSP006) |
|
推荐默认完全培养基货号 |
|
|
B方案完全培养基货号 |
|
|
生物安全等级 |
BSL-1 |
|
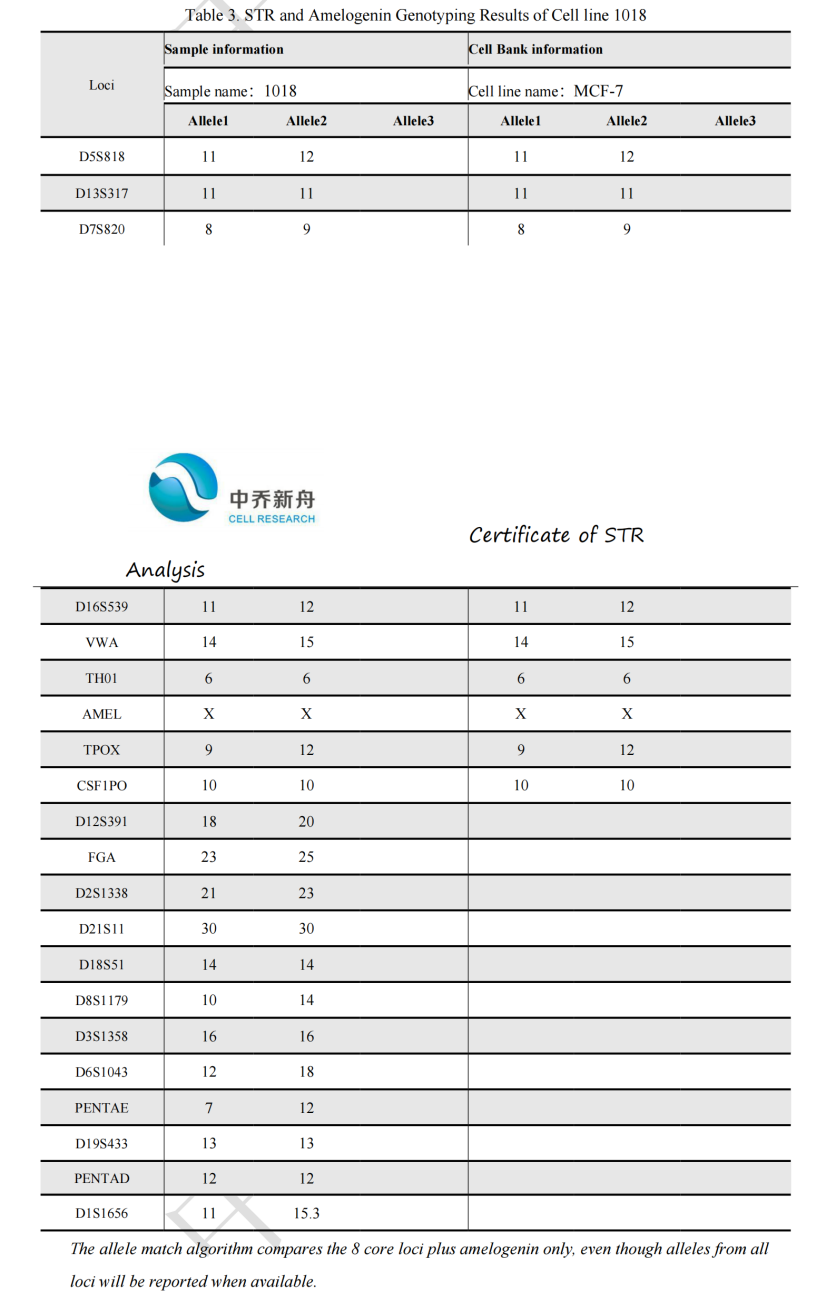
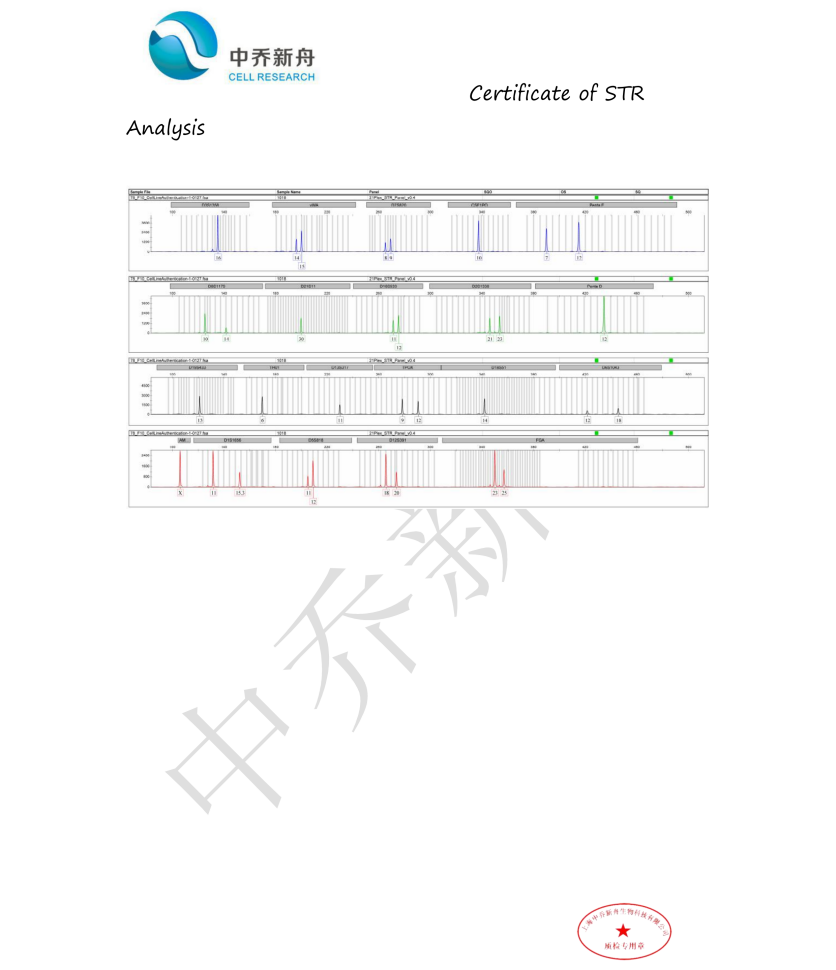
STR位点信息 |
Amelogenin: X
D3S1358: 16 |
|
培养条件 |
95%空气,5%二氧化碳;37℃ |
|
抗原表达/受体表达 |
*** |
|
基因表达 |
*** |
|
保藏机构 |
ATCC; CRL-12584 ATCC; HTB-22 BCRC; 60436 BCRJ; 0162 DSMZ; ACC-115 ECACC; 86012803 |
|
供应限制 |
仅供科研使用 |
|
货号 |
ZQ0071 |
|
发货规格 |
活细胞:T25培养瓶*1瓶或者1ml 冻存管*2支(细胞量约为1x10^6 Cells/Vial)二选一 |
|
发货形式 |
活细胞:常温运输;冻存管:干冰运输 |
|
储存温度 |
活细胞:培养箱;冻存管:液氮罐 |
|
产地 |
中国 |
|
供应限制 |
仅供科研使用 |
| 货号 | 产品名称 | 规格 | 价格 | 指令 |
 上海中乔新舟生物科技有限公司
上海中乔新舟生物科技有限公司