|
产品名称 |
HBL-100人整合SV40基因的乳腺上皮细胞 |
|
货号 |
ZQ0370 |
|
产品介绍 |
HBL-100是一种人类乳腺上皮细胞系,最初来源于哺乳期母亲的母乳。母乳是在分娩后三天收集的,尽管供体没有乳房病变的证据,也没有乳腺癌家族史,但细胞在第7代时表现出异常的核型。该细胞系以其合成少量乳糖的能力和通过增加酪蛋白的产生对催乳素或雌激素的刺激作出反应而闻名。显微镜分析,如电子显微镜,证实了这些细胞中存在微绒毛、张力原纤维和桥粒,突出了它们典型的上皮特征。
注意事项: |
|
种属 |
人 |
|
性别/年龄 |
女 |
|
组织 |
整合SV40基因的乳腺上皮细胞 |
|
细胞类型 |
转化细胞系 |
|
形态学 |
上皮 |
|
生长方式 |
贴壁 |
|
倍增时间 |
大约28~40小时 |
|
培养基和添加剂 |
RPMI-1640(中乔新舟 货号:ZQ-200)+20%胎牛血清(中乔新舟 货号:ZQ0500)+1%P/S(中乔新舟 货号:CSP006) |
|
推荐完全培养基货号 |
|
|
生物安全等级 |
BSL-1 |
|
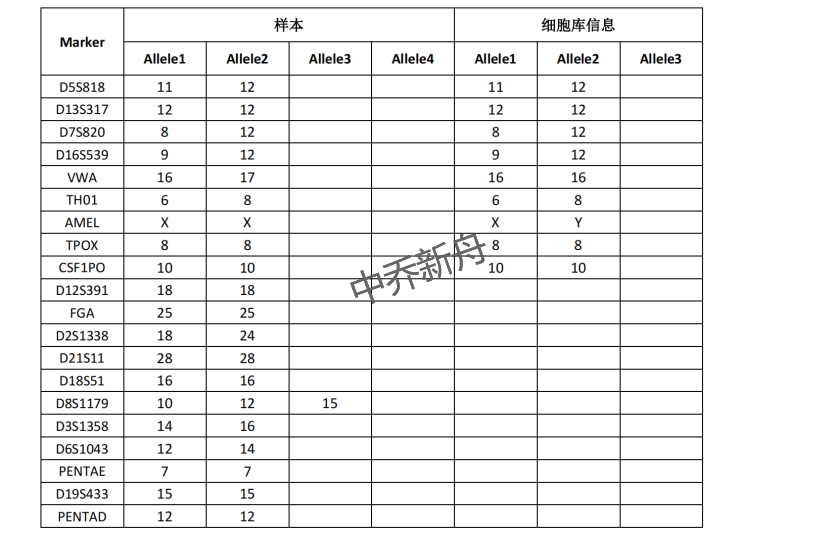
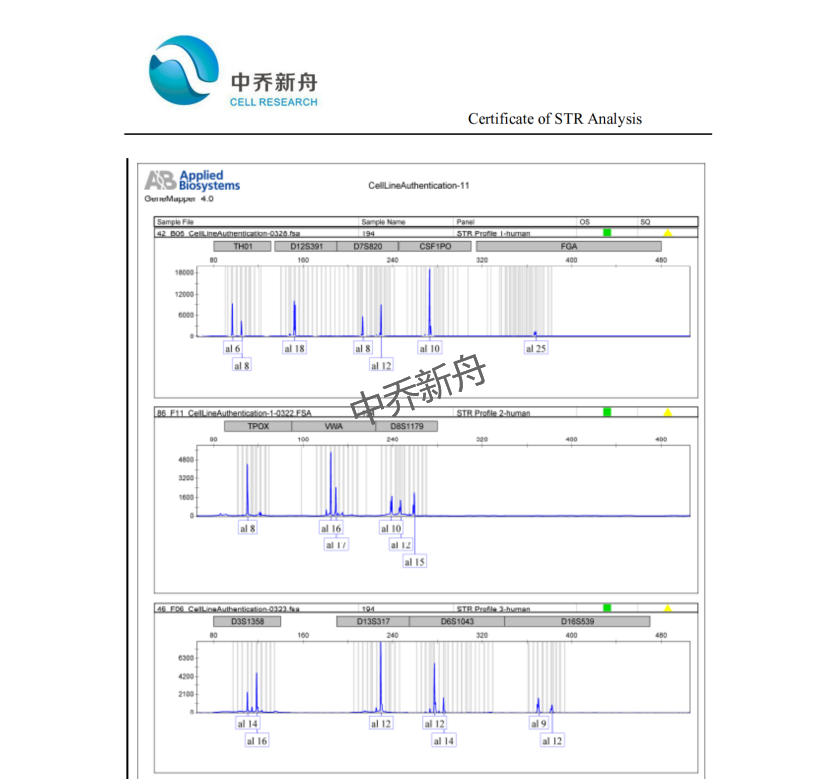
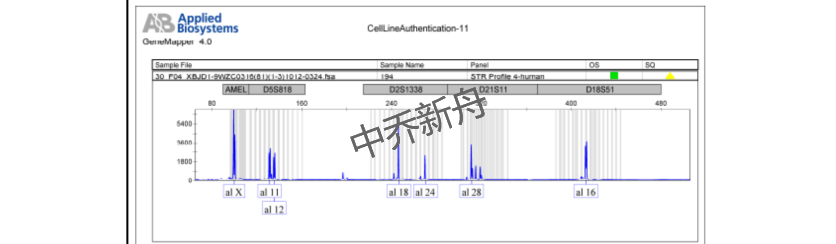
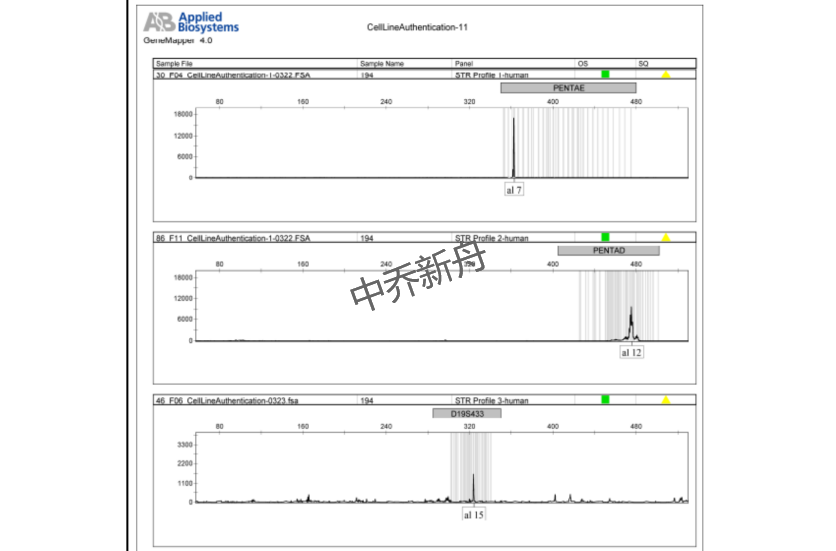
STR位点信息 |
Amelogenin: X |
|
培养条件 |
95%空气,5%二氧化碳;37℃ |
|
抗原表达/受体表达 |
*** |
|
基因表达 |
*** |
|
保藏机构 |
ATCC; HTB-124 RCB; RCB0460 |
|
供应限制 |
仅供科研使用 |
|
货号 |
ZQ0370 |
|
发货规格 |
活细胞:T25培养瓶*1瓶或者1ml 冻存管*2支(细胞量约为1x10^6 Cells/Vial)二选一 |
|
发货形式 |
活细胞:常温运输;冻存管:干冰运输 |
|
储存温度 |
活细胞:培养箱;冻存管:液氮罐 |
|
产地 |
中国 |
|
供应限制 |
仅供科研使用 |
| 货号 | 产品名称 | 规格 | 价格 | 指令 |
| ZQ0156 | COS-1非洲绿猴SV40转化的肾细胞(种属鉴定)[细胞+500ml专培套餐促销] | 1x10^6 Cells/Vial | ¥980.00 | 放入购物车 》 |
| ZQ0157 | COS-7非洲绿猴SV40转化的肾细胞(种属鉴定)[细胞+500ml专培套餐促销] | 1x10^6 Cells/Vial | ¥980.00 | 放入购物车 》 |
| ZQ0194 | SV40 MES 13小鼠肾小球系膜细胞(种属鉴定)[细胞+500ml专培套餐促销] | 1x10^6 Cells/Vial | ¥980.00 | 放入购物车 》 |
| ZQ0402 | hFOB 1.19人SV40转染成骨细胞(暂不销售) | 1x10^6 Cells/Vial | ¥1450.00 | 放入购物车 》 |
| ZQ505 | 支原体检测试剂盒(买二送一) | 1ml/100T | ¥800.00 | 放入购物车 》 |
| ZQ506 | 支原体清除试剂盒 | 1ml | ¥600.00 | 放入购物车 》 |
| A1001-1 | 国产转染试剂 | 1ml | ¥800.00 | 放入购物车 》 |
| 1 | 慢病毒介导基因沉默或过表达 | ¥询价 | 放入购物车 》 | |
| ZQ500 | 国产优级胎牛血清 | 500ml | ¥1800.00 | 放入购物车 》 |
| 5 | 荧光素酶标记 | ¥询价 | 放入购物车 》 | |
| 5 | 荧光素酶标记 | ¥询价 | 放入购物车 》 | |
| 5 | 荧光素酶标记 | ¥询价 | 放入购物车 》 | |
| ZQ-200 | RPMI-1640基础培养基 | 500ml | ¥56.00 | 放入购物车 》 |
| ZQ-201 | RPMI-1640完全培养基(10%FBS) | 500ml | ¥350.00 | 放入购物车 》 |
 上海中乔新舟生物科技有限公司
上海中乔新舟生物科技有限公司