
|
产品名称 |
MM.1S人IgA-骨髓瘤细胞 |
|
货号 |
ZQ0485 |
|
产品介绍 |
MM.1S是一种人IgA-骨髓瘤细胞系,最初是从患有免疫球蛋白Aλ骨髓瘤的42岁黑人女性患者的外周血中分离的B淋巴母细胞建立的,这些细胞具有淋巴母细胞样的形态,并且既可以悬浮生长也可以轻微贴壁。可用于研究多发性骨髓瘤的疾病进展、药物治疗反应以及耐药性机制。对地塞米松敏感。
注意事项: |
|
种属 |
人 |
|
性别/年龄 |
女/42岁 |
|
组织 |
外周血 |
|
疾病 |
免疫球蛋白Aλ骨髓瘤 |
|
细胞类型 |
B淋巴细胞 |
|
形态学 |
淋巴母细胞 |
|
生长方式 |
半悬浮 |
|
倍增时间 |
大约72~80小时 |
|
培养基和添加剂 |
RPMI-1640(中乔新舟 货号:ZQ-200)+10%胎牛血清(中乔新舟 货号:ZQ0500)+1%双抗(中乔新舟 货号:CSP006)+1% Sodium Pyruvate 100 mM Solution(中乔新舟 货号:CSP003) +1%L-alanyl-L-glutamine(中乔新舟 货号:CSP004) |
|
推荐完全培养基货号 |
|
|
生物安全等级 |
BSL-1 |
|
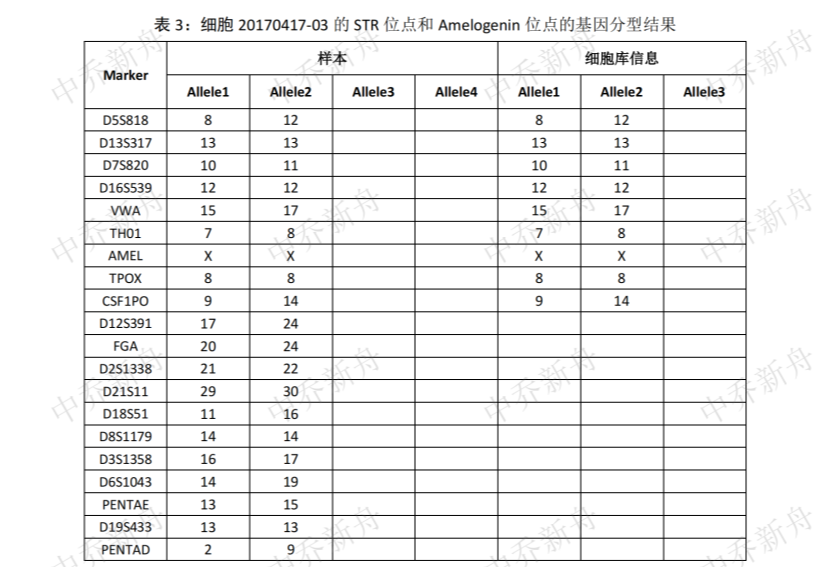
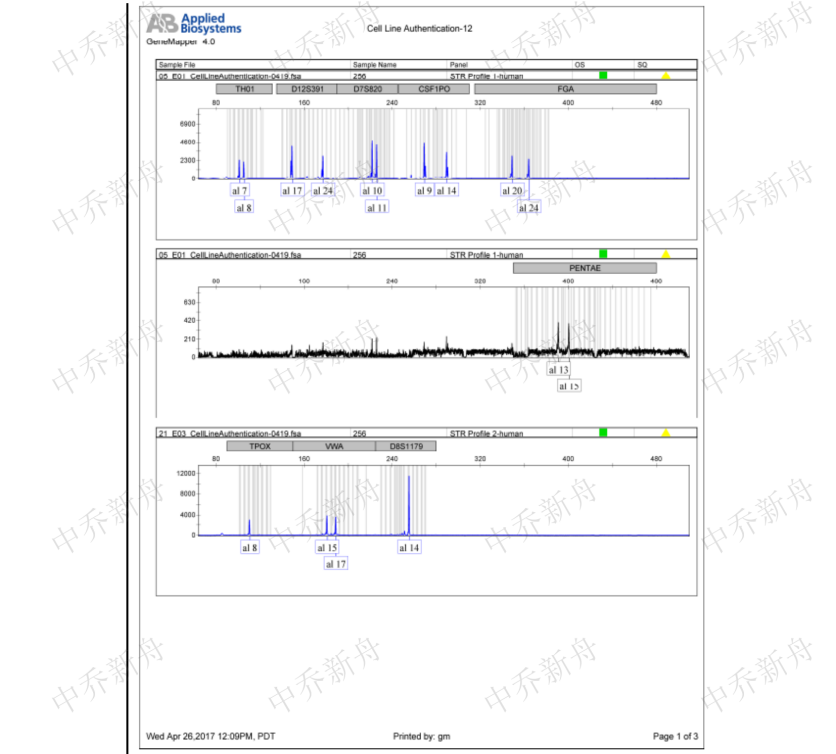
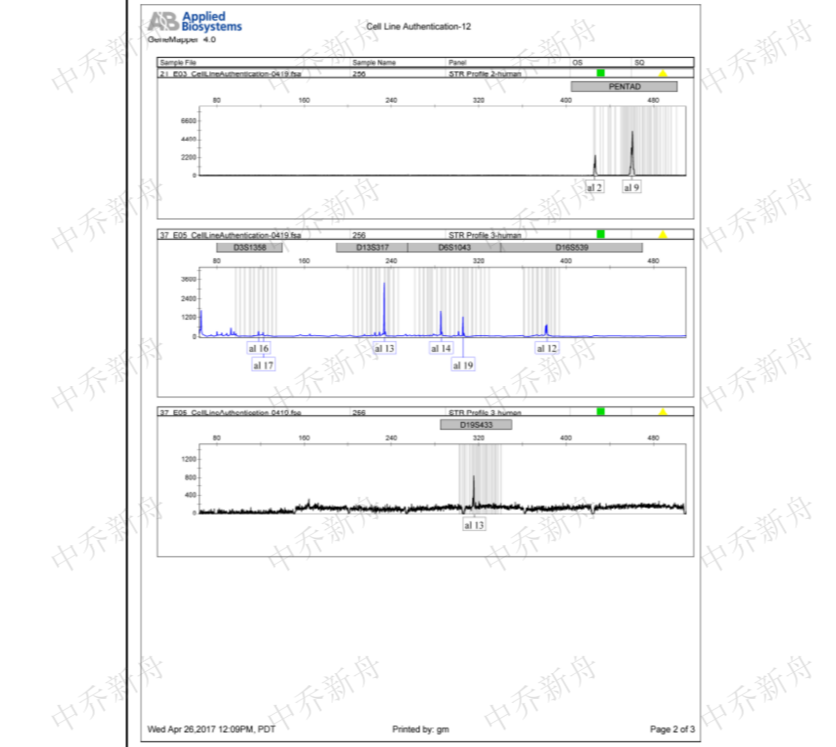
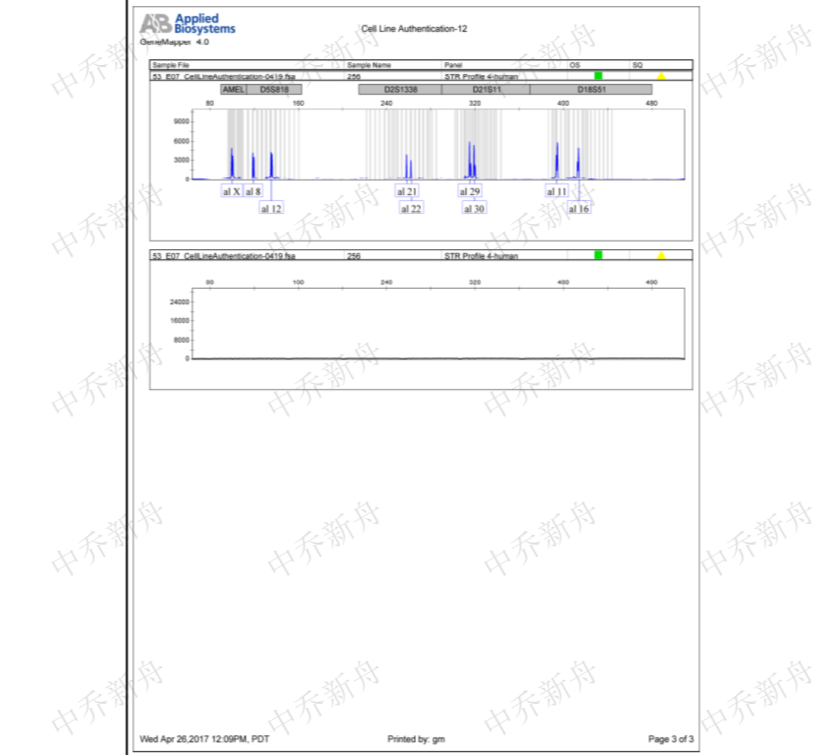
STR位点信息 |
Amelogenin: X
|
|
培养条件 |
95%空气,5%二氧化碳;37℃ |
|
抗原表达/受体表达 |
文献报道抗原表达CD25、CD38、CD52和CD59,糖皮质激素受体。 |
|
基因表达 |
*** |
|
保藏机构 |
ATCC; CRL-2974 |
|
供应限制 |
仅供科研使用 |
|
货号 |
ZQ0485 |
|
发货规格 |
活细胞:T25培养瓶*1瓶或者1ml 冻存管*2支(细胞量约为1x10^6 Cells/Vial)二选一 |
|
发货形式 |
活细胞:常温运输;冻存管:干冰运输 |
|
储存温度 |
活细胞:培养箱;冻存管:液氮罐 |
|
产地 |
中国 |
|
供应限制 |
仅供科研使用 |
原文链接: https://www.frontiersin.org/articles/10.3389/fimmu.2020.01292/full
PubMed=12691914; DOI=10.1016/S0301-472X(03)00023-7
Greenstein S., Krett N.L., Kurosawa Y., Ma C.-G., Chauhan D., Hideshima T., Anderson K.C., Rosen S.T.
Characterization of the MM.1 human multiple myeloma (MM) cell lines: a model system to elucidate the characteristics, behavior, and signaling of steroid-sensitive and -resistant MM cells.
Exp. Hematol. 31:271-282(2003)
PubMed=14760100; DOI=10.1158/1078-0432.CCR-0793-03
Shammas M.A., Shmookler Reis R.J., Li C., Koley H., Hurley L.H., Anderson K.C., Munshi N.C.
Telomerase inhibition and cell growth arrest after telomestatin treatment in multiple myeloma.
Clin. Cancer Res. 10:770-776(2004)
PubMed=16956823
Bataille R., Jego G., Robillard N., Barille-Nion S., Harousseau J.-L., Moreau P., Amiot M., Pellat-Deceunynck C.
The phenotype of normal, reactive and malignant plasma cells Identification of 'many and multiple myelomas' and of new targets for myeloma therapy.
Haematologica 91:1234-1240(2006)
PubMed=17692805; DOI=10.1016/j.ccr.2007.07.003; PMCID=PMC2083698
Keats J.J., Fonseca R., Chesi M., Schop R., Baker A., Chng W.-J., Van Wier S., Tiedemann R., Shi C.-X., Sebag M., Braggio E., Henry T., Zhu Y.-X., Fogle H., Price-Troska T.L., Ahmann G.J., Mancini C., Brents L.A., Kumar S.K., Greipp P.R., Dispenzieri A., Bryant B., Mulligan G., Bruhn L., Barrett M.T., Valdez R., Trent J.M., Stewart A.K., Carpten J.D., Bergsagel P.L.
Promiscuous mutations activate the noncanonical NF-kappaB pathway in multiple myeloma.
Cancer Cell 12:131-144(2007)
PubMed=18647998; DOI=10.1093/jncimonographs/lgn011; PMCID=PMC2737184
Dib A., Gabrea A., Glebov O.K., Bergsagel P.L., Kuehl W.M.
Characterization of MYC translocations in multiple myeloma cell lines.
J. Natl. Cancer Inst. Monogr. 39:25-31(2008)
PubMed=21173094; DOI=10.3324/haematol.2010.033456; PMCID=PMC3069235
Moreaux J., Klein B., Bataille R., Descamps G., Maiga S., Hose D., Goldschmidt H., Jauch A., Reme T., Jourdan M., Amiot M., Pellat-Deceunynck C.
A high-risk signature for patients with multiple myeloma established from the molecular classification of human myeloma cell lines.
Haematologica 96:574-582(2011)
PubMed=22460905; DOI=10.1038/nature11003; PMCID=PMC3320027
Barretina J.G., Caponigro G., Stransky N., Venkatesan K., Margolin A.A., Kim S., Wilson C.J., Lehar J., Kryukov G.V., Sonkin D., Reddy A., Liu M., Murray L., Berger M.F., Monahan J.E., Morais P., Meltzer J., Korejwa A., Jane-Valbuena J., Mapa F.A., Thibault J., Bric-Furlong E., Raman P., Shipway A., Engels I.H., Cheng J., Yu G.-Y.K., Yu J.-J., Aspesi P. Jr., de Silva M., Jagtap K., Jones M.D., Wang L., Hatton C., Palescandolo E., Gupta S., Mahan S., Sougnez C., Onofrio R.C., Liefeld T., MacConaill L.E., Winckler W., Reich M., Li N.-X., Mesirov J.P., Gabriel S.B., Getz G., Ardlie K., Chan V., Myer V.E., Weber B.L., Porter J., Warmuth M., Finan P., Harris J.L., Meyerson M.L., Golub T.R., Morrissey M.P., Sellers W.R., Schlegel R., Garraway L.A.
The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity.
Nature 483:603-607(2012)
PubMed=25984343; DOI=10.1038/sdata.2014.35; PMCID=PMC4432652
Cowley G.S., Weir B.A., Vazquez F., Tamayo P., Scott J.A., Rusin S., East-Seletsky A., Ali L.D., Gerath W.F.J., Pantel S.E., Lizotte P.H., Jiang G.-Z., Hsiao J., Tsherniak A., Dwinell E., Aoyama S., Okamoto M., Harrington W., Gelfand E.T., Green T.M., Tomko M.J., Gopal S., Wong T.C., Li H.-B., Howell S., Stransky N., Liefeld T., Jang D., Bistline J., Meyers B.H., Armstrong S.A., Anderson K.C., Stegmaier K., Reich M., Pellman D., Boehm J.S., Mesirov J.P., Golub T.R., Root D.E., Hahn W.C.
Parallel genome-scale loss of function screens in 216 cancer cell lines for the identification of context-specific genetic dependencies.
Sci. Data 1:140035-140035(2014)
PubMed=25485619; DOI=10.1038/nbt.3080
Klijn C., Durinck S., Stawiski E.W., Haverty P.M., Jiang Z.-S., Liu H.-B., Degenhardt J., Mayba O., Gnad F., Liu J.-F., Pau G., Reeder J., Cao Y., Mukhyala K., Selvaraj S.K., Yu M.-M., Zynda G.J., Brauer M.J., Wu T.D., Gentleman R.C., Manning G., Yauch R.L., Bourgon R., Stokoe D., Modrusan Z., Neve R.M., de Sauvage F.J., Settleman J., Seshagiri S., Zhang Z.-M.
A comprehensive transcriptional portrait of human cancer cell lines.
Nat. Biotechnol. 33:306-312(2015)
PubMed=25688540; DOI=10.1002/cyto.a.22643
Maiga S., Brosseau C., Descamps G., Dousset C., Gomez-Bougie P., Chiron D., Menoret E., Kervoelen C., Vie H., Cesbron A., Moreau-Aubry A., Amiot M., Pellat-Deceunynck C.
A simple flow cytometry-based barcode for routine authentication of multiple myeloma and mantle cell lymphoma cell lines.
Cytometry A 87:285-288(2015)
PubMed=25877200; DOI=10.1038/nature14397
Yu M., Selvaraj S.K., Liang-Chu M.M.Y., Aghajani S., Busse M., Yuan J., Lee G., Peale F.V., Klijn C., Bourgon R., Kaminker J.S., Neve R.M.
A resource for cell line authentication, annotation and quality control.
Nature 520:307-311(2015)
PubMed=26589293; DOI=10.1186/s13073-015-0240-5; PMCID=PMC4653878
Scholtalbers J., Boegel S., Bukur T., Byl M., Goerges S., Sorn P., Loewer M., Sahin U., Castle J.C.
TCLP: an online cancer cell line catalogue integrating HLA type, predicted neo-epitopes, virus and gene expression.
Genome Med. 7:118.1-118.7(2015)
PubMed=27397505; DOI=10.1016/j.cell.2016.06.017; PMCID=PMC4967469
Iorio F., Knijnenburg T.A., Vis D.J., Bignell G.R., Menden M.P., Schubert M., Aben N., Goncalves E., Barthorpe S., Lightfoot H., Cokelaer T., Greninger P., van Dyk E., Chang H., de Silva H., Heyn H., Deng X.-M., Egan R.K., Liu Q.-S., Mironenko T., Mitropoulos X., Richardson L., Wang J.-H., Zhang T.-H., Moran S., Sayols S., Soleimani M., Tamborero D., Lopez-Bigas N., Ross-Macdonald P., Esteller M., Gray N.S., Haber D.A., Stratton M.R., Benes C.H., Wessels L.F.A., Saez-Rodriguez J., McDermott U., Garnett M.J.
A landscape of pharmacogenomic interactions in cancer.
Cell 166:740-754(2016)
PubMed=28196595; DOI=10.1016/j.ccell.2017.01.005; PMCID=PMC5501076
Li J., Zhao W., Akbani R., Liu W.-B., Ju Z.-L., Ling S.-Y., Vellano C.P., Roebuck P., Yu Q.-H., Eterovic A.K., Byers L.A., Davies M.A., Deng W.-L., Gopal Y.N.V., Chen G., von Euw E.M., Slamon D.J., Conklin D., Heymach J.V., Gazdar A.F., Minna J.D., Myers J.N., Lu Y.-L., Mills G.B., Liang H.
Characterization of human cancer cell lines by reverse-phase protein arrays.
Cancer Cell 31:225-239(2017)
PubMed=30285677; DOI=10.1186/s12885-018-4840-5; PMCID=PMC6167786
Tan K.-T., Ding L.-W., Sun Q.-Y., Lao Z.-T., Chien W., Ren X., Xiao J.-F., Loh X.-Y., Xu L., Lill M., Mayakonda A., Lin D.-C., Yang H.H., Koeffler H.P.
Profiling the B/T cell receptor repertoire of lymphocyte derived cell lines.
BMC Cancer 18:940.1-940.13(2018)
PubMed=30545397; DOI=10.1186/s13045-018-0679-0; PMCID=PMC6293660
Tessoulin B., Moreau-Aubry A., Descamps G., Gomez-Bougie P., Maiga S., Gaignard A., Chiron D., Menoret E., Le Gouill S., Moreau P., Amiot M., Pellat-Deceunynck C.
Whole-exon sequencing of human myeloma cell lines shows mutations related to myeloma patients at relapse with major hits in the DNA regulation and repair pathways.
J. Hematol. Oncol. 11:137.1-137.13(2018)
PubMed=30894373; DOI=10.1158/0008-5472.CAN-18-2747; PMCID=PMC6445675
Dutil J., Chen Z.-H., Monteiro A.N.A., Teer J.K., Eschrich S.A.
An interactive resource to probe genetic diversity and estimated ancestry in cancer cell lines.
Cancer Res. 79:1263-1273(2019)
PubMed=30971826; DOI=10.1038/s41586-019-1103-9
Behan F.M., Iorio F., Picco G., Goncalves E., Beaver C.M., Migliardi G., Santos R., Rao Y., Sassi F., Pinnelli M., Ansari R., Harper S., Jackson D.A., McRae R., Pooley R., Wilkinson P., van der Meer D.J., Dow D., Buser-Doepner C.A., Bertotti A., Trusolino L., Stronach E.A., Saez-Rodriguez J., Yusa K., Garnett M.J.
Prioritization of cancer therapeutic targets using CRISPR-Cas9 screens.
Nature 568:511-516(2019)
PubMed=31068700; DOI=10.1038/s41586-019-1186-3; PMCID=PMC6697103
Ghandi M., Huang F.W., Jane-Valbuena J., Kryukov G.V., Lo C.C., McDonald E.R. 3rd, Barretina J.G., Gelfand E.T., Bielski C.M., Li H.-X., Hu K., Andreev-Drakhlin A.Y., Kim J., Hess J.M., Haas B.J., Aguet F., Weir B.A., Rothberg M.V., Paolella B.R., Lawrence M.S., Akbani R., Lu Y.-L., Tiv H.L., Gokhale P.C., de Weck A., Mansour A.A., Oh C., Shih J., Hadi K., Rosen Y., Bistline J., Venkatesan K., Reddy A., Sonkin D., Liu M., Lehar J., Korn J.M., Porter D.A., Jones M.D., Golji J., Caponigro G., Taylor J.E., Dunning C.M., Creech A.L., Warren A.C., McFarland J.M., Zamanighomi M., Kauffmann A., Stransky N., Imielinski M., Maruvka Y.E., Cherniack A.D., Tsherniak A., Vazquez F., Jaffe J.D., Lane A.A., Weinstock D.M., Johannessen C.M., Morrissey M.P., Stegmeier F., Schlegel R., Hahn W.C., Getz G., Mills G.B., Boehm J.S., Golub T.R., Garraway L.A., Sellers W.R.
Next-generation characterization of the Cancer Cell Line Encyclopedia.
Nature 569:503-508(2019)
PubMed=32123307; DOI=10.1038/s41375-020-0785-1; PMCID=PMC7483300
Sarin V., Yu K., Ferguson I.D., Gugliemini O., Nix M.A., Hann B., Sirota M., Wiita A.P.
Evaluating the efficacy of multiple myeloma cell lines as models for patient tumors via transcriptomic correlation analysis.
Leukemia 34:2754-2765(2020)
PubMed=35839778; DOI=10.1016/j.ccell.2022.06.010; PMCID=PMC9387775
Goncalves E., Poulos R.C., Cai Z.-X., Barthorpe S., Manda S.S., Lucas N., Beck A., Bucio-Noble D., Dausmann M., Hall C., Hecker M., Koh J., Lightfoot H., Mahboob S., Mali I., Morris J., Richardson L., Seneviratne A.J., Shepherd R., Sykes E., Thomas F., Valentini S., Williams S.G., Wu Y.-X., Xavier D., MacKenzie K.L., Hains P.G., Tully B., Robinson P.J., Zhong Q., Garnett M.J., Reddel R.R.
Pan-cancer proteomic map of 949 human cell lines.
Cancer Cell 40:835-849.e8(2022)
| 产品名称 | 价格 | 指令 |
| Y3-Ag 1.2.3大鼠骨髓瘤细胞(种属鉴定) | ¥1800.00 | 购物车 》 |
| NCI-H929人骨髓瘤细胞株 (STR鉴定) | ¥1800.00 | 购物车 》 |
| FO小鼠骨髓瘤细胞(种属鉴定) | ¥1350.00 | 购物车 》 |
| P3/NSI/1-Ag4-1 [NS-1]小鼠骨髓瘤细胞(种属鉴定)[细胞+500ml专培套餐促销] | ¥980.00 | 购物车 》 |
| P3X63Ag8小鼠骨髓瘤细胞(种属鉴定)[细胞+500ml专培套餐促销] | ¥980.00 | 购物车 》 |
| P3X63Ag8.653小鼠骨髓瘤细胞(种属鉴定) | ¥1000.00 | 购物车 》 |
| U266B1人骨髓瘤细胞 | ¥1500.00 | 购物车 》 |
 上海中乔新舟生物科技有限公司
上海中乔新舟生物科技有限公司