|
产品名称 |
KYSE-30 人食管鳞癌细胞 |
|
货号 |
ZQ0963 |
|
产品介绍 |
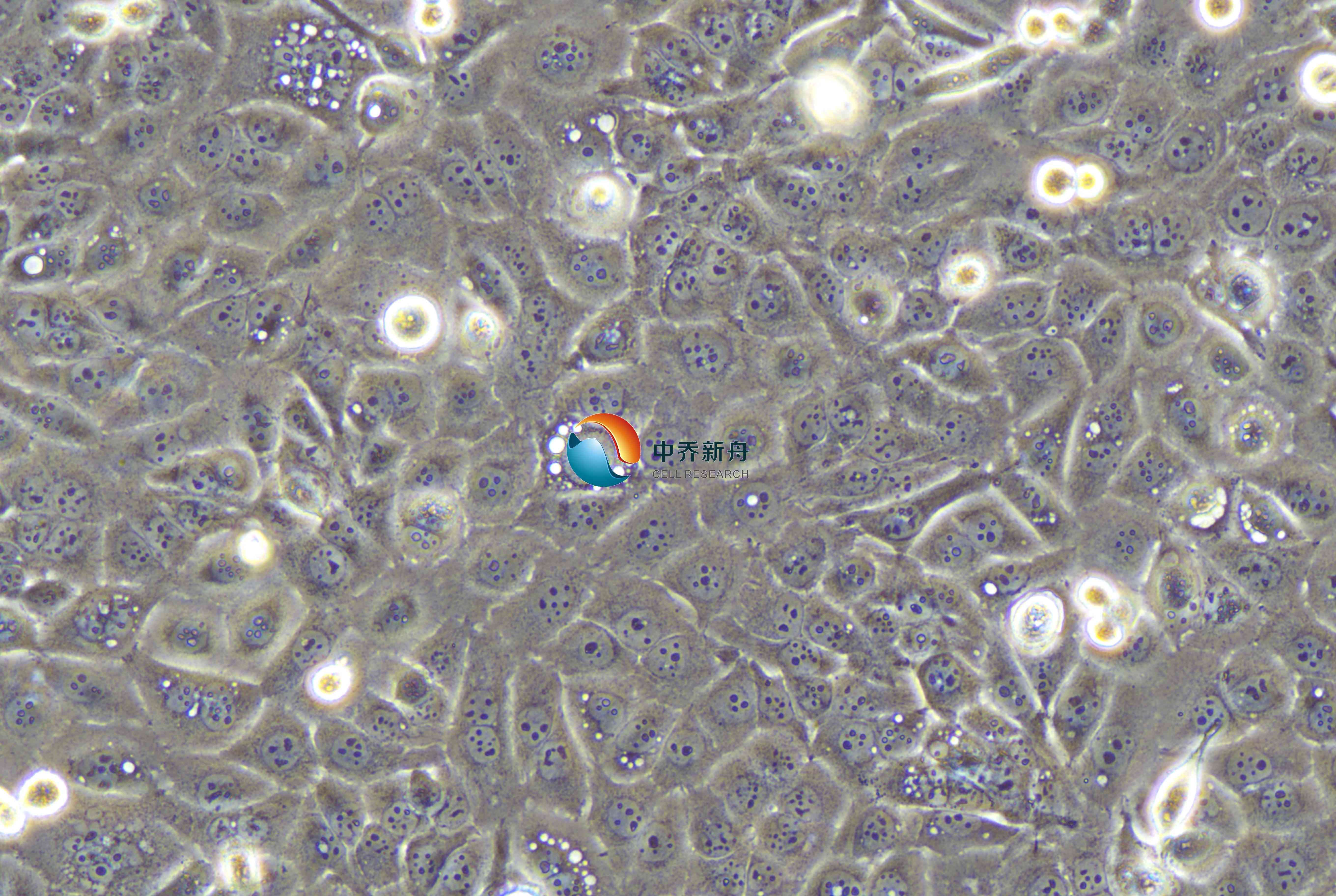
这株细胞来源于治疗前从一名64岁日本男性胸中段食管切除的高分化侵袭性食管鳞状细胞癌;将肿瘤细胞异种移植到无胸腺小鼠中建立细胞系;在文献中描述为可在无胸腺小鼠中异源移植,并携带p53突变和cERB、MYC和CYCLIN D1的扩增。作为KYSE系列的一部分,该细胞系的建立是为了研究食管癌的分子和细胞特征。该细胞主要生长为贴壁单层,在相差显微镜下显示出典型的多边形形状和均匀的外观。它的生长模式是典型的上皮来源的癌细胞,形成紧密排列的集落,并倾向于以无序的方式堆积,反映了它们来源于肿瘤的侵袭性。
从遗传学上讲,KYSE-30在关键肿瘤抑制基因的改变上具有重要意义。该细胞系显示出p16 (INK4a)和p15 (INK4b)基因的野生型配置,但它在p16基因中携带一个显著的点突变,导致过早停止密码子,导致截断,无功能蛋白。这种突变可能导致细胞周期控制的丧失,促进癌细胞不受控制的增殖特征。然而,野生型p15基因的保留表明,p16基因的改变在KYSE-30的肿瘤发生中起着更关键的作用,这可能与关注这些基因在癌症中的差异作用的研究有关。 |
|
种属 |
人 |
|
性别/年龄 |
男/64岁 |
|
组织 |
食管鳞状上皮 |
|
疾病 |
食管鳞状细胞癌 |
|
细胞类型 |
肿瘤细胞 |
|
形态学 |
上皮细胞样,带有长的伪足 |
|
生长方式 |
贴壁 |
|
倍增时间 |
20.8 hours (PubMed=1728357); 22 hours (PubMed=25984343); ~30 hours (DSMZ=ACC-351) |
|
培养基和添加剂 |
RPMI-1640(中乔新舟 货号:ZQ-200)+F-12(中乔新舟 货号:ZQ-400)+2mM L-Glutamine(中乔新舟 货号:CSP0012)+10%胎牛血清(中乔新舟 货号:ZQ0500)+1%双抗(中乔新舟 货号:CSP006) |
|
推荐完全培养基货号 |
|
|
生物安全等级 |
BSL-1 |
|
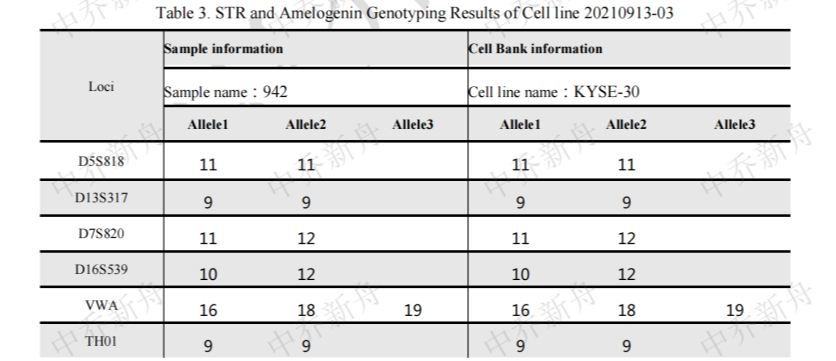
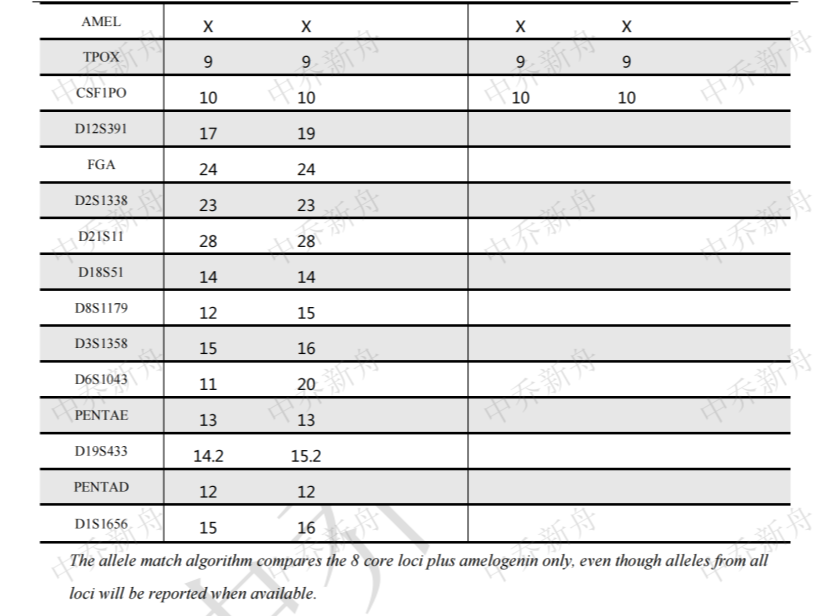
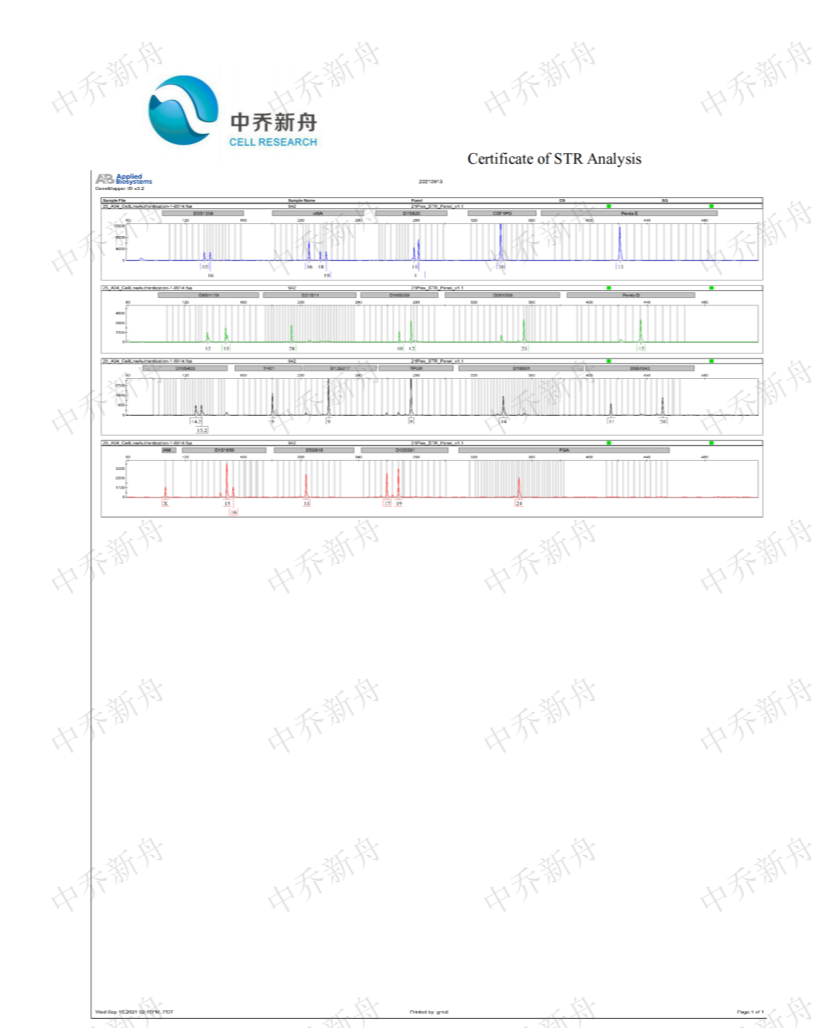
STR位点信息 |
Amelogenin: X CSF1PO: 10 D2S1338: 23 D3S1358: 15,16 D5S818: 11 D7S820: 11,11.3 (JCRB=JCRB0188; PubMed=25877200) :11,12 (DSMZ=ACC-351) D8S1179: 12,15 D13S317: 9 D16S539: 10,12 D18S51: 14 D19S433: 15,16 D21S11: 28 FGA: 24 Penta D: 12 Penta E: 13 TH01: 9 TPOX: 8,9 (JCRB=JCRB0188) :9 (DSMZ=ACC-351; PubMed=25877200) vWA: 16,18 (DSMZ=ACC-351) :16,18,19 (JCRB=JCRB0188; PubMed=25877200) |
|
培养条件 |
95%空气,5%二氧化碳;37℃ |
|
抗原表达/受体表达 |
*** |
|
基因表达 |
*** |
|
保藏机构 |
DSMZ; ACC-351 ECACC; 94072011 JCRB; JCRB0188 |
|
供应限制 |
仅供科研使用 |
|
货号 |
ZQ0963 |
|
发货规格 |
活细胞:T25培养瓶*1瓶或者1ml 冻存管*2支(细胞量约为1x10^6 Cells/Vial )二选一 |
|
发货形式 |
活细胞:常温运输;冻存管:干冰运输 |
|
储存温度 |
活细胞:培养箱;冻存管:液氮罐 |
|
产地 |
中国 |
|
供应限制 |
仅供科研使用 |
原文链接: https://academic.oup.com/abbs/article-abstract/52/8/842/5869218
PubMed=1728357; DOI=10.1002/1097-0142(19920115)69:2<277::AID-CNCR2820690202>3.0.CO;2-C
Shimada Y., Imamura M., Wagata T., Yamaguchi N., Tobe T.
Characterization of 21 newly established esophageal cancer cell lines.
Cancer 69:277-284(1992)
PubMed=7913084; DOI=10.1002/ijc.2910580224
Kanda Y., Nishiyama Y., Shimada Y., Imamura M., Nomura H., Hiai H., Fukumoto M.
Analysis of gene amplification and overexpression in human esophageal-carcinoma cell lines.
Int. J. Cancer 58:291-297(1994)
PubMed=8575860; DOI=10.1002/(SICI)1097-0215(19960126)65:3<372::AID-IJC16>3.0.CO;2-C
Tanaka H., Shibagaki I., Shimada Y., Wagata T., Imamura M., Ishizaki K.
Characterization of p53 gene mutations in esophageal squamous cell carcinoma cell lines: increased frequency and different spectrum of mutations from primary tumors.
Int. J. Cancer 65:372-376(1996)
PubMed=9033652; DOI=10.1002/(SICI)1097-0215(19970207)70:4<437::AID-IJC11>3.0.CO;2-C
Tanaka H., Shimada Y., Imamura M., Shibagaki I., Ishizaki K.
Multiple types of aberrations in the p16 (INK4a) and the p15(INK4b) genes in 30 esophageal squamous-cell-carcinoma cell lines.
Int. J. Cancer 70:437-442(1997)
PubMed=11092977; DOI=10.1111/j.1349-7006.2000.tb00895.x
Pimkhaokham A., Shimada Y., Fukuda Y., Kurihara N., Imoto I., Yang Z.-Q., Imamura M., Nakamura Y., Amagasa T., Inazawa J.
Nonrandom chromosomal imbalances in esophageal squamous cell carcinoma cell lines: possible involvement of the ATF3 and CENPF genes in the 1q32 amplicon.
Jpn. J. Cancer Res. 91:1126-1133(2000)
PubMed=11520067; DOI=10.1006/bbrc.2001.5400
Kan T., Shimada Y., Sato F., Maeda M., Kawabe A., Kaganoi J.-i., Itami A., Yamasaki S., Imamura M.
Gene expression profiling in human esophageal cancers using cDNA microarray.
Biochem. Biophys. Res. Commun. 286:792-801(2001)
PubMed=12963126; DOI=10.1016/S0304-3835(03)00344-6
Hoque M.O., Begum S., Sommer M., Lee T., Trink B., Ratovitski E., Sidransky D.
PUMA in head and neck cancer.
Cancer Lett. 199:75-81(2003)
PubMed=15172977; DOI=10.1158/0008-5472.CAN-04-0172
Sonoda I., Imoto I., Inoue J., Shibata T., Shimada Y., Chin K., Imamura M., Amagasa T., Gray J.W., Hirohashi S., Inazawa J.
Frequent silencing of low density lipoprotein receptor-related protein 1B (LRP1B) expression by genetic and epigenetic mechanisms in esophageal squamous cell carcinoma.
Cancer Res. 64:3741-3747(2004)
PubMed=16045545; DOI=10.1111/j.0959-9673.2005.00431.x
Ban S., Michikawa Y., Ishikawa K.-i., Sagara M., Watanabe K., Shimada Y., Inazawa J., Imai T.
Radiation sensitivities of 31 human oesophageal squamous cell carcinoma cell lines.
Int. J. Exp. Pathol. 86:231-240(2005)
PubMed=16364037; DOI=10.1111/j.1442-2050.2006.00530.x
Su M., Chin S.-F., Li X.-Y., Edwards P.A.W., Caldas C., Fitzgerald R.C.
Comparative genomic hybridization of esophageal adenocarcinoma and squamous cell carcinoma cell lines.
Dis. Esophagus 19:10-14(2006)
PubMed=20215515; DOI=10.1158/0008-5472.CAN-09-3458
Rothenberg S.M., Mohapatra G., Rivera M.N., Winokur D., Greninger P., Nitta M., Sadow P.M., Sooriyakumar G., Brannigan B.W., Ulman M.J., Perera R.M., Wang R., Tam A., Ma X.-J., Erlander M., Sgroi D.C., Rocco J.W., Lingen M.W., Cohen E.E.W., Louis D.N., Settleman J., Haber D.A.
A genome-wide screen for microdeletions reveals disruption of polarity complex genes in diverse human cancers.
Cancer Res. 70:2158-2164(2010)
PubMed=21191746; DOI=10.1007/s11684-010-0260-x
Ji J.-F., Wu K., Wu M., Zhan Q.-M.
p53 functional activation is independent of its genotype in five esophageal squamous cell carcinoma cell lines.
Front. Med. China 4:412-418(2010)
PubMed=22460905; DOI=10.1038/nature11003
Barretina J.G., Caponigro G., Stransky N., Venkatesan K., Margolin A.A., Kim S., Wilson C.J., Lehar J., Kryukov G.V., Sonkin D., Reddy A., Liu M., Murray L., Berger M.F., Monahan J.E., Morais P., Meltzer J., Korejwa A., Jane-Valbuena J., Mapa F.A., Thibault J., Bric-Furlong E., Raman P., Shipway A., Engels I.H., Cheng J., Yu G.-Y.K., Yu J.-J., Aspesi P. Jr., de Silva M., Jagtap K., Jones M.D., Wang L., Hatton C., Palescandolo E., Gupta S., Mahan S., Sougnez C., Onofrio R.C., Liefeld T., MacConaill L.E., Winckler W., Reich M., Li N.-X., Mesirov J.P., Gabriel S.B., Getz G., Ardlie K., Chan V., Myer V.E., Weber B.L., Porter J., Warmuth M., Finan P., Harris J.L., Meyerson M.L., Golub T.R., Morrissey M.P., Sellers W.R., Schlegel R., Garraway L.A.
The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity.
Nature 483:603-607(2012)
PubMed=25984343; DOI=10.1038/sdata.2014.35
Cowley G.S., Weir B.A., Vazquez F., Tamayo P., Scott J.A., Rusin S., East-Seletsky A., Ali L.D., Gerath W.F.J., Pantel S.E., Lizotte P.H., Jiang G.-Z., Hsiao J., Tsherniak A., Dwinell E., Aoyama S., Okamoto M., Harrington W., Gelfand E.T., Green T.M., Tomko M.J., Gopal S., Wong T.C., Li H.-B., Howell S., Stransky N., Liefeld T., Jang D., Bistline J., Meyers B.H., Armstrong S.A., Anderson K.C., Stegmaier K., Reich M., Pellman D., Boehm J.S., Mesirov J.P., Golub T.R., Root D.E., Hahn W.C.
Parallel genome-scale loss of function screens in 216 cancer cell lines for the identification of context-specific genetic dependencies.
Sci. Data 1:140035-140035(2014)
 上海中乔新舟生物科技有限公司
上海中乔新舟生物科技有限公司