|
产品名称 |
Mino人非霍奇金淋巴瘤细胞 |
|
货号 |
ZQ0995 |
|
产品介绍 |
Mino人非霍奇金淋巴瘤细胞(Mino Human non-Hodgkin Lymphoma Cell Line)是一种从人类患者身上获取并经过实验室培养的细胞系,用于科学研究。这个特定的细胞系是从一位64岁男性的外周血中提取的,与非霍奇金淋巴瘤(NHL)相关。非霍奇金淋巴瘤是一种发生在淋巴系统的癌症,涉及淋巴细胞的异常增殖。
这些细胞很大,在体外单独生长并形成小团块。流式细胞仪检测的免疫表型与 MCL 一致。蛋白质印迹显示细胞周期蛋白 D1 表达,但未检测到细胞周期蛋白 D2 和细胞周期蛋白 D3;视网膜母细胞瘤蛋白主要被磷酸化。表达抑癌基因产物,包括 p53、p16 (INK4a) 和 p21 (WAF1)。 细胞很大,单独生长,在体外成小块生长。 |
|
种属 |
人 |
|
性别/年龄 |
男/64岁 |
|
组织 |
外周血 |
|
疾病 |
套细胞淋巴瘤 |
|
生物安全等级 |
BSL-1 |
|
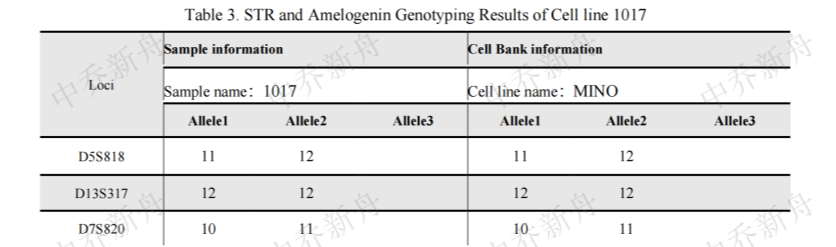
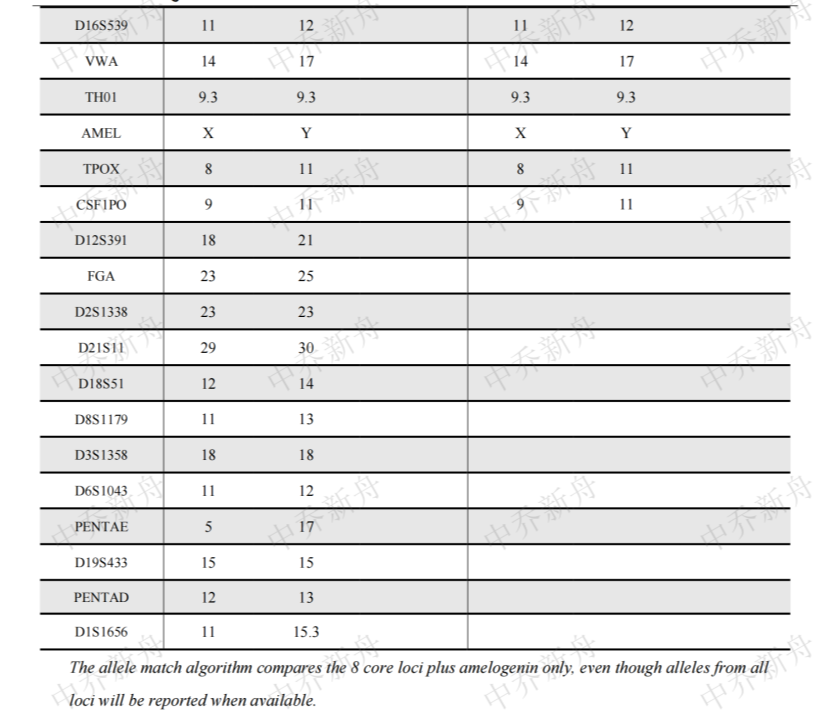
STR位点信息 |
Amelogenin: X,Y
CSF1PO:9,11
|
|
细胞类型 |
成淋巴细胞 |
|
形态学 |
淋巴母细胞 |
|
生长方式 |
悬浮 |
|
倍增时间 |
72 hours (PubMed=25315077); ~50 hours (DSMZ=ACC-687) |
|
培养基和添加剂 |
RPMI 1640 (中乔新舟 货号:ZQ-200)+15%FBS(中乔新舟 货号:ZQ0500)+1%PS(中乔新舟 货号:CSP006) |
|
推荐完全培养基货号 |
|
|
培养条件 |
95%空气,5%二氧化碳;37℃ |
|
抗原表达/受体表达 |
*** |
|
基因表达 |
*** |
|
保藏机构 |
ATCC; CRL-3000 DSMZ; ACC-687 |
|
供应限制 |
仅供科研使用 |
|
货号 |
ZQ0995 |
|
发货规格 |
活细胞:T25培养瓶*1瓶或者1ml 冻存管*2支(细胞量约为1x10^6 Cells/Vial )二选一 |
|
发货形式 |
活细胞:常温运输;冻存管:干冰运输 |
|
储存温度 |
活细胞:培养箱;冻存管:液氮罐 |
|
产地 |
中国 |
|
供应限制 |
仅供科研使用 |
PubMed=12127561; DOI=10.1016/S0145-2126(02)00013-9
Lai R., McDonnell T.J., O'Connor S.L., Medeiros L.J., Oudat R.I., Keating M.J., Morgan M.B., Curiel T.J., Ford R.J. Jr.
Establishment and characterization of a new mantle cell lymphoma cell line, Mino.
Leuk. Res. 26:849-855(2002)
PubMed=12683869; DOI=10.5858/2003-127-0424-COMCLC
Amin H.M., McDonnell T.J., Medeiros L.J., Rassidakis G.Z., Leventaki V., O'Connor S.L., Keating M.J., Lai R.
Characterization of 4 mantle cell lymphoma cell lines. Establishment of an in vitro study model.
Arch. Pathol. Lab. Med. 127:424-431(2003)
PubMed=21746927; DOI=10.1073/pnas.1018941108
Beltran E., Fresquet V., Martinez-Useros J., Richter-Larrea J.A., Sagardoy A., Sesma I., Almada L.L., Montes-Moreno S., Siebert R., Gesk S., Calasanz M.J., Malumbres R., Rieger M., Prosper F., Lossos I.S., Piris M.A., Fernandez-Zapico M.E., Martinez-Climent J.A.
A cyclin-D1 interaction with BAX underlies its oncogenic role and potential as a therapeutic target in mantle cell lymphoma.
Proc. Natl. Acad. Sci. U.S.A. 108:12461-12466(2011)
PubMed=22460905; DOI=10.1038/nature11003
Barretina J.G., Caponigro G., Stransky N., Venkatesan K., Margolin A.A., Kim S., Wilson C.J., Lehar J., Kryukov G.V., Sonkin D., Reddy A., Liu M., Murray L., Berger M.F., Monahan J.E., Morais P., Meltzer J., Korejwa A., Jane-Valbuena J., Mapa F.A., Thibault J., Bric-Furlong E., Raman P., Shipway A., Engels I.H., Cheng J., Yu G.-Y.K., Yu J.-J., Aspesi P. Jr., de Silva M., Jagtap K., Jones M.D., Wang L., Hatton C., Palescandolo E., Gupta S., Mahan S., Sougnez C., Onofrio R.C., Liefeld T., MacConaill L.E., Winckler W., Reich M., Li N.-X., Mesirov J.P., Gabriel S.B., Getz G., Ardlie K., Chan V., Myer V.E., Weber B.L., Porter J., Warmuth M., Finan P., Harris J.L., Meyerson M.L., Golub T.R., Morrissey M.P., Sellers W.R., Schlegel R., Garraway L.A.
The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity.
Nature 483:603-607(2012)
PubMed=24362935; DOI=10.1038/nm.3435
Rahal R., Frick M., Romero R., Korn J.M., Kridel R., Chan F.C., Meissner B., Bhang H.-E.C., Ruddy D., Kauffmann A., Farsidjani A., Derti A., Rakiec D., Naylor T., Pfister E., Kovats S., Kim S., Dietze K., Dorken B., Steidl C., Tzankov A., Hummel M., Monahan J., Morrissey M.P., Fritsch C., Sellers W.R., Cooke V.G., Gascoyne R.D., Lenz G., Stegmeier F.
Pharmacological and genomic profiling identifies NF-kappaB-targeted treatment strategies for mantle cell lymphoma.
Nat. Med. 20:87-92(2014)
PubMed=25960936; DOI=10.4161/21624011.2014.954893
Boegel S., Lower M., Bukur T., Sahin U., Castle J.C.
A catalog of HLA type, HLA expression, and neo-epitope candidates in human cancer cell lines.
OncoImmunology 3:e954893.1-e954893.12(2014)
PubMed=25315077; DOI=10.3109/10428194.2014.970548
Fogli L.K., Williams M.E., Connors J.M., Reid Y.A., Brown K., O'Connor O.A.
Development and characterization of a Mantle Cell Lymphoma Cell Bank in the American Type Culture Collection.
Leuk. Lymphoma 56:2114-2122(2015)
PubMed=25355872; DOI=10.1128/JVI.02570-14
Cao S.-B., Strong M.J., Wang X., Moss W.N., Concha M., Lin Z., O'Grady T., Baddoo M., Fewell C., Renne R., Flemington E.K.
High-throughput RNA sequencing-based virome analysis of 50 lymphoma cell lines from the Cancer Cell Line Encyclopedia project.
J. Virol. 89:713-729(2015)
PubMed=25688540; DOI=10.1002/cyto.a.22643
Maiga S., Brosseau C., Descamps G., Dousset C., Gomez-Bougie P., Chiron D., Menoret E., Kervoelen C., Vie H., Cesbron A., Moreau-Aubry A., Amiot M., Pellat-Deceunynck C.
A simple flow cytometry-based barcode for routine authentication of multiple myeloma and mantle cell lymphoma cell lines.
Cytometry A 87:285-288(2015)
PubMed=26194763; DOI=10.1182/blood-2015-01-624585
Bhatt S., Matthews J., Parvin S., Sarosiek K.A., Zhao D.-K., Jiang X.-Y., Isik E., Letai A.G., Lossos I.S.
Direct and immune-mediated cytotoxicity of interleukin-21 contributes to antitumor effects in mantle cell lymphoma.
Blood 126:1555-1564(2015)
PubMed=28196595; DOI=10.1016/j.ccell.2017.01.005
Li J., Zhao W., Akbani R., Liu W.-B., Ju Z.-L., Ling S.-Y., Vellano C.P., Roebuck P., Yu Q.-H., Eterovic A.K., Byers L.A., Davies M.A., Deng W.-L., Gopal Y.N.V., Chen G., von Euw E.M., Slamon D.J., Conklin D., Heymach J.V., Gazdar A.F., Minna J.D., Myers J.N., Lu Y.-L., Mills G.B., Liang H.
Characterization of human cancer cell lines by reverse-phase protein arrays.
Cancer Cell 31:225-239(2017)
PubMed=29666304; DOI=10.1158/1078-0432.CCR-17-3004
Pham L.V., Huang S.-J., Zhang H., Zhang J., Bell T., Zhou S.-H., Pogue E., Ding Z.-Y., Lam L., Westin J., Davis R.E., Young K.H., Medeiros L.J., Ford R.J. Jr., Nomie K., Zhang L., Wang M.
Strategic therapeutic targeting to overcome venetoclax resistance in aggressive B-cell lymphomas.
Clin. Cancer Res. 24:3967-3980(2018)
PubMed=30285677; DOI=10.1186/s12885-018-4840-5
Tan K.-T., Ding L.-W., Sun Q.-Y., Lao Z.-T., Chien W., Ren X., Xiao J.-F., Loh X.-Y., Xu L., Lill M., Mayakonda A., Lin D.-C., Yang H., Koeffler H.P.
Profiling the B/T cell receptor repertoire of lymphocyte derived cell lines.
BMC Cancer 18:940.1-940.13(2018)
PubMed=30894373; DOI=10.1158/0008-5472.CAN-18-2747
Dutil J., Chen Z.-H., Monteiro A.N.A., Teer J.K., Eschrich S.A.
An interactive resource to probe genetic diversity and estimated ancestry in cancer cell lines.
Cancer Res. 79:1263-1273(2019)
PubMed=31068700; DOI=10.1038/s41586-019-1186-3
Ghandi M., Huang F.W., Jane-Valbuena J., Kryukov G.V., Lo C.C., McDonald E.R. III, Barretina J.G., Gelfand E.T., Bielski C.M., Li H.-X., Hu K., Andreev-Drakhlin A.Y., Kim J., Hess J.M., Haas B.J., Aguet F., Weir B.A., Rothberg M.V., Paolella B.R., Lawrence M.S., Akbani R., Lu Y.-L., Tiv H.L., Gokhale P.C., de Weck A., Mansour A.A., Oh C., Shih J., Hadi K., Rosen Y., Bistline J., Venkatesan K., Reddy A., Sonkin D., Liu M., Lehar J., Korn J.M., Porter D.A., Jones M.D., Golji J., Caponigro G., Taylor J.E., Dunning C.M., Creech A.L., Warren A.C., McFarland J.M., Zamanighomi M., Kauffmann A., Stransky N., Imielinski M., Maruvka Y.E., Cherniack A.D., Tsherniak A., Vazquez F., Jaffe J.D., Lane A.A., Weinstock D.M., Johannessen C.M., Morrissey M.P., Stegmeier F., Schlegel R., Hahn W.C., Getz G., Mills G.B., Boehm J.S., Golub T.R., Garraway L.A., Sellers W.R.
Next-generation characterization of the Cancer Cell Line Encyclopedia.
Nature 569:503-508(2019)
PubMed=31160637; DOI=10.1038/s41598-019-44491-x
Quentmeier H., Pommerenke C., Dirks W.G., Eberth S., Koeppel M., MacLeod R.A.F., Nagel S., Steube K., Uphoff C.C., Drexler H.G.
The LL-100 panel: 100 cell lines for blood cancer studies.
Sci. Rep. 9:8218-8218(2019)

 上海中乔新舟生物科技有限公司
上海中乔新舟生物科技有限公司