|
产品名称 |
SK-MEL-28人恶性黑色素瘤细胞 |
|
货号 |
ZQ0997 |
|
产品说明 |
SK-MEL-28是一种人类恶性黑色素瘤细胞系,最初由Takahashi T及其同事从一位51岁男性患者的皮肤组织中分离建立。这种细胞系具有亚四倍体的核型,模态染色体数为90,并且具有复杂的核型和多个染色体异常。该细胞系是合适的转染宿主,在癌症和毒理学研究中具有应用。 |
|
种属 |
人 |
|
性别/年龄 |
男性/51岁 |
|
组织 |
皮肤 |
|
疾病 |
恶性黑色素瘤 |
|
生物安全等级 |
BSL-1 |
|
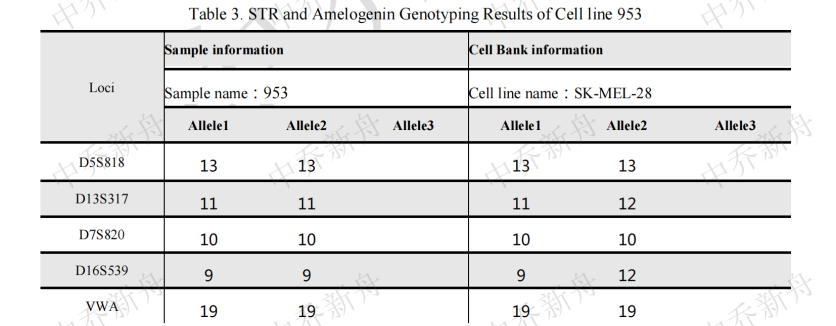
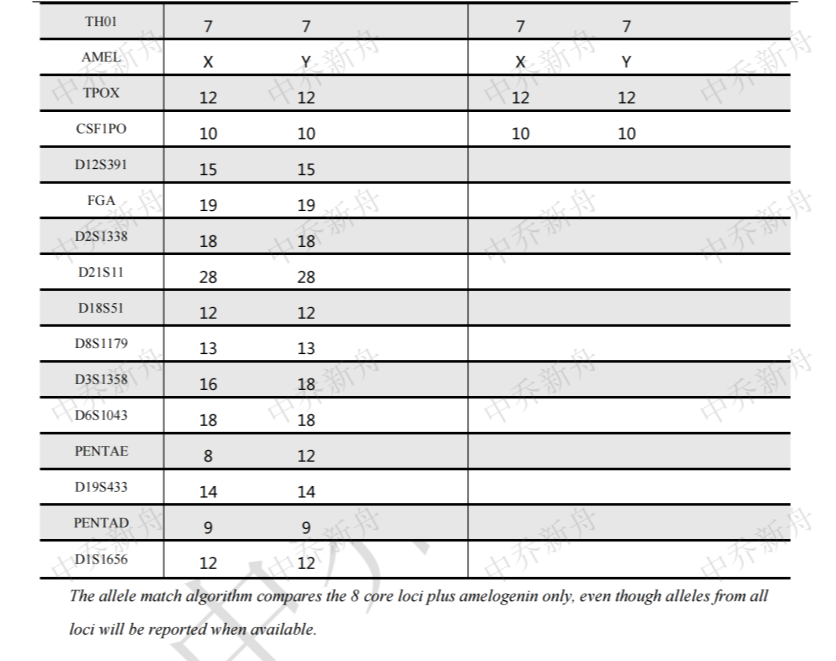
STR位点信息 |
Amelogenin: X,Y D3S1358: 16,18 TH01: 7 D21S11: 28,29 D18S51: 12,16 Penta_E: 8,12 D5S818: 11,13 D13S317: 11,12 D7S820: 9.3,10 D16S539: 9,12 CSF1PO: 10,12 Penta_D: 9,10 vWA: 16,19 D8S1179: 13 TPOX: 8,12 FGA: 19 D19S433: 14 D2S1338: 18vWA: 16,19 |
|
细胞类型 |
肿瘤细胞 |
|
形态学 |
多边形 |
|
生长方式 |
贴壁生长 |
|
倍增时间 |
27 hours (PubMed=7718330); 72 hours (PubMed=12479222); 26.9 hours (PubMed=20406486); 35.1 hours (NCI-DTP=SK-MEL-28) |
|
培养基和添加剂 |
MEM(中乔新舟 货号:ZQ-300)+10%FBS(中乔新舟 货号:ZQ500-A)+1%PS(中乔新舟 货号:CSP006) |
|
推荐完全培养基货号 |
|
|
培养条件 |
95%空气,5%二氧化碳;37℃ |
|
抗原表达/受体表达 |
*** |
|
基因表达 |
*** |
|
保藏机构 |
ATCC; HTB-72 BCRJ; 0289 |
|
供应限制 |
仅供科研使用 |
|
货号 |
ZQ0997 |
|
发货规格 |
活细胞:T25培养瓶*1瓶或者1ml 冻存管*2支(细胞量约为1x10^6 Cells/Vial )二选一 |
|
发货形式 |
活细胞:常温运输;冻存管:干冰运输 |
|
储存温度 |
活细胞:培养箱;冻存管:液氮罐 |
|
产地 |
中国 |
|
供应限制 |
仅供科研使用 |
PubMed=978138; DOI=10.1084/jem.144.4.873
Shiku H., Takahashi T., Oettgen H.F., Old L.J.
Cell surface antigens of human malignant melanoma. II. Serological typing with immune adherence assays and definition of two new surface antigens.
J. Exp. Med. 144:873-881(1976)
PubMed=1067619; DOI=10.1073/pnas.73.9.3278
Carey T.E., Takahashi T., Resnick-Silverman L.A., Oettgen H.F., Old L.J.
Cell surface antigens of human malignant melanoma: mixed hemadsorption assays for humoral immunity to cultured autologous melanoma cells.
Proc. Natl. Acad. Sci. U.S.A. 73:3278-3282(1976)
PubMed=327080; DOI=10.1093/jnci/59.1.221
Fogh J., Fogh J.M., Orfeo T.
One hundred and twenty-seven cultured human tumor cell lines producing tumors in nude mice.
J. Natl. Cancer Inst. 59:221-226(1977)
PubMed=313568; DOI=10.1073/pnas.76.6.2898
Carey T.E., Lloyd K.O., Takahashi T., Travassos L.R., Old L.J.
AU cell-surface antigen of human malignant melanoma: solubilization and partial characterization.
Proc. Natl. Acad. Sci. U.S.A. 76:2898-2902(1979)
PubMed=6933476; DOI=10.1073/pnas.77.7.4260
Houghton A.N., Taormina M.C., Ikeda H., Watanabe T., Oettgen H.F., Old L.J.
Serological survey of normal humans for natural antibody to cell surface antigens of melanoma.
Proc. Natl. Acad. Sci. U.S.A. 77:4260-4264(1980)
DOI=10.1007/978-1-4615-7228-2_39
Houghton A.N., Oettgen H.F., Old L.J.
Malignant melanoma. Current status of the search for melanoma-specific antigens.
(In) Immunodermatology. Comprehensive Immunology, Vol 7; Safai B., Good R.A. (eds.); pp.557-576; Springer; Boston (1981)
PubMed=7017212; DOI=10.1093/jnci/66.6.1003
Pollack M.S., Heagney S.D., Livingston P.O., Fogh J.
HLA-A, B, C and DR alloantigen expression on forty-six cultured human tumor cell lines.
J. Natl. Cancer Inst. 66:1003-1012(1981)
PubMed=7175440; DOI=10.1084/jem.156.6.1755
Houghton A.N., Eisinger M., Albino A.P., Cairncross J.G., Old L.J.
Surface antigens of melanocytes and melanomas. Markers of melanocyte differentiation and melanoma subsets.
J. Exp. Med. 156:1755-1766(1982)
PubMed=6197381; DOI=10.1002/ijc.2910320610
Mattes M.J., Thomson T.M., Old L.J., Lloyd K.O.
A pigmentation-associated, differentiation antigen of human melanoma defined by a precipitating antibody in human serum.
Int. J. Cancer 32:717-721(1983)
PubMed=6349783; DOI=10.1002/1097-0142(19830915)52:6<949::AID-CNCR2820520602>3.0.CO;2-2
Dantas M.E., Brown J.P., Thomas M.R., Robinson W.A., Glode L.M.
Detection of melanoma cells in bone marrow using monoclonal antibodies. A comparison of fluorescence activated cell sorting (FACS) and conventional immunofluorescence (IF).
Cancer 52:949-953(1983)
PubMed=6864164; DOI=10.1084/jem.158.1.53
Houghton A.N., Brooks H., Cote R.J., Taormina M.C., Oettgen H.F., Old L.J.
Detection of cell surface and intracellular antigens by human monoclonal antibodies. Hybrid cell lines derived from lymphocytes of patients with malignant melanoma.
J. Exp. Med. 158:53-65(1983)
PubMed=6203018; DOI=10.1111/j.1348-0421.1984.tb00673.x
Maezawa N., Yano A.
Two distinct cytotoxic T lymphocyte subpopulations in patients with Vogt-Koyanagi-Harada disease that recognize human melanoma cells.
Microbiol. Immunol. 28:219-231(1984)
PubMed=6582512; DOI=10.1073/pnas.81.2.568
Mattes M.J., Cordon-Cardo C., Lewis J.L. Jr., Old L.J., Lloyd K.O.
Cell surface antigens of human ovarian and endometrial carcinoma defined by mouse monoclonal antibodies.
Proc. Natl. Acad. Sci. U.S.A. 81:568-572(1984)
PubMed=2983346; DOI=10.1073/pnas.82.5.1470
Dracopoli N.C., Houghton A.N., Old L.J.
Loss of polymorphic restriction fragments in malignant melanoma: implications for tumor heterogeneity.
Proc. Natl. Acad. Sci. U.S.A. 82:1470-1474(1985)
Patent=US4591572
Mattes M.J., Thomson T.M., Old L.J., Lloyd K.O.
Pigmentation associated, differentiation antigen of human melanoma and autologous antibody.
Patent number US4591572, 27-May-1986
PubMed=3518877; DOI=10.3109/07357908609038260
Fogh J.
Human tumor lines for cancer research.
Cancer Invest. 4:157-184(1986)
PubMed=3335022
Alley M.C., Scudiero D.A., Monks A., Hursey M.L., Czerwinski M.J., Fine D.L., Abbott B.J., Mayo J.G., Shoemaker R.H., Boyd M.R.
Feasibility of drug screening with panels of human tumor cell lines using a microculture tetrazolium assay.
Cancer Res. 48:589-601(1988)
PubMed=1716514
Albino A.P., Davis B.M., Nanus D.M.
Induction of growth factor RNA expression in human malignant melanoma: markers of transformation.
Cancer Res. 51:4815-4820(1991)
PubMed=2041050; DOI=10.1093/jnci/83.11.757
Monks A., Scudiero D.A., Skehan P., Shoemaker R.H., Paull K.D., Vistica D.T., Hose C.D., Langley J., Cronise P., Vaigro-Wolff A., Gray-Goodrich M., Campbell H., Mayo J.G., Boyd M.R.
Feasibility of a high-flux anticancer drug screen using a diverse panel of cultured human tumor cell lines.
J. Natl. Cancer Inst. 83:757-766(1991)
PubMed=2068080; DOI=10.1073/pnas.88.14.6028
Cornil I., Theodorescu D., Man S., Herlyn M., Jambrosic J.A., Kerbel R.S.
Fibroblast cell interactions with human melanoma cells affect tumor cell growth as a function of tumor progression.
Proc. Natl. Acad. Sci. U.S.A. 88:6028-6032(1991)
PubMed=7999427; DOI=10.1016/0959-8049(94)90188-0
Marshall E.S., Matthews J.H.L., Shaw J.H.F., Nixon J., Tumewu P., Finlay G.J., Holdaway K.M., Baguley B.C.
Radiosensitivity of new and established human melanoma cell lines: comparison of [3H]thymidine incorporation and soft agar clonogenic assays.
Eur. J. Cancer 30:1370-1376(1994)
PubMed=7718330; DOI=10.1016/0959-8049(94)00472-H
Baguley B.C., Marshall E.S., Whittaker J.R., Dotchin M.C., Nixon J., McCrystal M.R., Finlay G.J., Matthews J.H.L., Holdaway K.M., van Zijl P.L.
Resistance mechanisms determining the in vitro sensitivity to paclitaxel of tumour cells cultured from patients with ovarian cancer.
Eur. J. Cancer 31:230-237(1995)
PubMed=9354451
Castellano M., Pollock P.M., Walters M.K., Sparrow L.E., Down L.M., Gabrielli B.G., Parsons P.G., Hayward N.K.
CDKN2A/p16 is inactivated in most melanoma cell lines.
Cancer Res. 57:4868-4875(1997)
PubMed=9466661; DOI=10.1002/(SICI)1097-0215(19980209)75:4<590::AID-IJC16>3.0.CO;2-D
White C.A., Thomson S.A., Cooper L.T., van Endert P.M., Tampe R., Coupar B., Qiu L., Parsons P.G., Moss D.J., Khanna R.
Constitutive transduction of peptide transporter and HLA genes restores antigen processing function and cytotoxic T cell-mediated immune recognition of human melanoma cells.
Int. J. Cancer 75:590-595(1998)
PubMed=9598804; DOI=10.1002/(SICI)1098-2264(199806)22:2<157::AID-GCC11>3.0.CO;2-N
Walker G.J., Flores J.F., Glendening J.M., Lin A.H.-T., Markl I.D.C., Fountain J.W.
Virtually 100% of melanoma cell lines harbor alterations at the DNA level within CDKN2A, CDKN2B, or one of their downstream targets.
Genes Chromosomes Cancer 22:157-163(1998)
PubMed=10700174; DOI=10.1038/73432
Ross D.T., Scherf U., Eisen M.B., Perou C.M., Rees C., Spellman P.T., Iyer V.R., Jeffrey S.S., van de Rijn M., Waltham M.C., Pergamenschikov A., Lee J.C.F., Lashkari D., Shalon D., Myers T.G., Weinstein J.N., Botstein D., Brown P.O.
Systematic variation in gene expression patterns in human cancer cell lines.
Nat. Genet. 24:227-235(2000)
PubMed=10766161
Tsao H., Zhang X., Fowlkes K., Haluska F.G.
Relative reciprocity of NRAS and PTEN/MMAC1 alterations in cutaneous melanoma cell lines.
Cancer Res. 60:1800-1804(2000)
PubMed=11016658
Girnita L., Girnita A., Brodin B., Xie Y.-T., Nilsson G., Dricu A., Lundeberg J., Wejde J., Bartolazzi A., Wiman K.G., Larsson O.
Increased expression of insulin-like growth factor I receptor in malignant cells expressing aberrant p53: functional impact.
Cancer Res. 60:5278-5283(2000)
PubMed=11121133; DOI=10.1046/j.1523-1747.2000.00199.x
Lacal P.M., Failla C.M., Pagani E., Odorisio T., Schietroma C., Falcinelli S., Zambruno G., D'Atri S.
Human melanoma cells secrete and respond to placenta growth factor and vascular endothelial growth factor.
J. Invest. Dermatol. 115:1000-1007(2000)
PubMed=12068308; DOI=10.1038/nature00766
Davies H., Bignell G.R., Cox C., Stephens P.J., Edkins S., Clegg S., Teague J.W., Woffendin H., Garnett M.J., Bottomley W., Davis N., Dicks E., Ewing R., Floyd Y., Gray K., Hall S., Hawes R., Hughes J., Kosmidou V., Menzies A., Mould C., Parker A., Stevens C., Watt S., Hooper S., Wilson R., Jayatilake H., Gusterson B.A., Cooper C.S., Shipley J.M., Hargrave D., Pritchard-Jones K., Maitland N.J., Chenevix-Trench G., Riggins G.J., Bigner D.D., Palmieri G., Cossu A., Flanagan A.M., Nicholson A., Ho J.W.C., Leung S.Y., Yuen S.T., Weber B.L., Seigler H.F., Darrow T.L., Paterson H.F., Marais R., Marshall C.J., Wooster R., Stratton M.R., Futreal P.A.
Mutations of the BRAF gene in human cancer.
Nature 417:949-954(2002)
PubMed=12479222
Wahl M.L., Owen J.A., Burd R., Herlands R.A., Nogami S.S., Rodeck U., Berd D., Leeper D.B., Owen C.S.
Regulation of intracellular pH in human melanoma: potential therapeutic implications.
Mol. Cancer Ther. 1:617-628(2002)
PubMed=14692828; DOI=10.1290/1543-706X(2004)40<35:COHMCL>2.0.CO;2
Quinones L.G., Garcia-Castro I.
Characterization of human melanoma cell lines according to their migratory properties in vitro.
In Vitro Cell. Dev. Biol. Anim. 40:35-42(2004)
PubMed=14871852; DOI=10.1158/0008-5472.CAN-03-2209
Hogan K.T., Coppola M.A., Gatlin C.L., Thompson L.W., Shabanowitz J., Hunt D.F., Engelhard V.H., Ross M.M., Slingluff C.L. Jr.
Identification of novel and widely expressed cancer/testis gene isoforms that elicit spontaneous cytotoxic T-lymphocyte reactivity to melanoma.
Cancer Res. 64:1157-1163(2004)
PubMed=15009714; DOI=10.1046/j.0022-202X.2004.22243.x
Tsao H., Goel V., Wu H., Yang G., Haluska F.G.
Genetic interaction between NRAS and BRAF mutations and PTEN/MMAC1 inactivation in melanoma.
J. Invest. Dermatol. 122:337-341(2004)
PubMed=15299072; DOI=10.1158/1535-7163.895.3.8
Qin J.-Z., Stennett L., Bacon P., Bodner B., Hendrix M.J.C., Seftor R.E.B., Seftor E.A., Margaryan N.V., Pollock P.M., Curtis A., Trent J.M., Bennett F., Miele L., Nickoloff B.J.
p53-independent NOXA induction overcomes apoptotic resistance of malignant melanomas.
Mol. Cancer Ther. 3:895-902(2004)
PubMed=15467732; DOI=10.1038/sj.onc.1208152
Tanami H., Imoto I., Hirasawa A., Yuki Y., Sonoda I., Inoue J., Yasui K., Misawa-Furihata A., Kawakami Y., Inazawa J.
Involvement of overexpressed wild-type BRAF in the growth of malignant melanoma cell lines.
Oncogene 23:8796-8804(2004)
PubMed=15748285; DOI=10.1186/1479-5876-3-11
Adams S., Robbins F.-M., Chen D., Wagage D., Holbeck S.L., Morse H.C. III, Stroncek D., Marincola F.M.
HLA class I and II genotype of the NCI-60 cell lines.
J. Transl. Med. 3:11.1-11.8(2005)
PubMed=17088437; DOI=10.1158/1535-7163.MCT-06-0433
Ikediobi O.N., Davies H., Bignell G.R., Edkins S., Stevens C., O'Meara S., Santarius T., Avis T., Barthorpe S., Brackenbury L., Buck G., Butler A.P., Clements J., Cole J., Dicks E., Forbes S., Gray K., Halliday K., Harrison R., Hills K., Hinton J., Hunter C., Jenkinson A., Jones D., Kosmidou V., Lugg R., Menzies A., Mironenko T., Parker A., Perry J., Raine K.M., Richardson D., Shepherd R., Small A., Smith R., Solomon H., Stephens P.J., Teague J.W., Tofts C., Varian J., Webb T., West S., Widaa S., Yates A., Reinhold W.C., Weinstein J.N., Stratton M.R., Futreal P.A., Wooster R.
Mutation analysis of 24 known cancer genes in the NCI-60 cell line set.
Mol. Cancer Ther. 5:2606-2612(2006)
PubMed=17260012; DOI=10.1038/sj.onc.1210252
Jonsson G., Dahl C., Staaf J., Sandberg T., Bendahl P.-O., Ringner M., Guldberg P., Borg A.
Genomic profiling of malignant melanoma using tiling-resolution arrayCGH.
Oncogene 26:4738-4748(2007)
PubMed=17308088; DOI=10.1158/0008-5472.CAN-06-3311
Shields J.M., Thomas N.E., Cregger M., Berger A.J., Leslie M., Torrice C., Hao H.-L., Penland S., Arbiser J.L., Scott G.A., Zhou T., Bar-Eli M., Bear J.E., Der C.J., Kaufmann W.K., Rimm D.L., Sharpless N.E.
Lack of extracellular signal-regulated kinase mitogen-activated protein kinase signaling shows a new type of melanoma.
Cancer Res. 67:1502-1512(2007)
PubMed=17363583; DOI=10.1158/0008-5472.CAN-06-4152
Stark M.S., Hayward N.K.
Genome-wide loss of heterozygosity and copy number analysis in melanoma using high-density single-nucleotide polymorphism arrays.
Cancer Res. 67:2632-2642(2007)
PubMed=17516929; DOI=10.1111/j.1600-0749.2007.00375.x
Johansson P., Pavey S., Hayward N.K.
Confirmation of a BRAF mutation-associated gene expression signature in melanoma.
Pigment Cell Res. 20:216-221(2007)
PubMed=18277095; DOI=10.4161/cbt.7.5.5712
Berglind H., Pawitan Y., Kato S., Ishioka C., Soussi T.
Analysis of p53 mutation status in human cancer cell lines: a paradigm for cell line cross-contamination.
Cancer Biol. Ther. 7:699-708(2008)
PubMed=18790768; DOI=10.1158/1535-7163.MCT-08-0431
Smalley K.S.M., Lioni M., Dalla Palma M., Xiao M., Desai B., Egyhazi Brage S., Hansson J., Wu H., King A.J., Van Belle P., Elder D.E., Flaherty K.T., Herlyn M., Nathanson K.L.
Increased cyclin D1 expression can mediate BRAF inhibitor resistance in BRAF V600E-mutated melanomas.
Mol. Cancer Ther. 7:2876-2883(2008)
PubMed=19147755; DOI=10.1158/1078-0432.CCR-08-1916
Augustine C.K., Yoo J.S., Potti A., Yoshimoto Y., Zipfel P.A., Friedman H.S., Nevins J.R., Ali-Osman F., Tyler D.S.
Genomic and molecular profiling predicts response to temozolomide in melanoma.
Clin. Cancer Res. 15:502-510(2009)
PubMed=19372543; DOI=10.1158/1535-7163.MCT-08-0921
Lorenzi P.L., Reinhold W.C., Varma S., Hutchinson A.A., Pommier Y., Chanock S.J., Weinstein J.N.
DNA fingerprinting of the NCI-60 cell line panel.
Mol. Cancer Ther. 8:713-724(2009)
PubMed=19799798; DOI=10.1186/1471-2407-9-352
Casula M., Muggiano A., Cossu A., Budroni M., Caraco C., Ascierto P.A., Pagani E., Stanganelli I., Canzanella S., Sini M.C., Palomba G., Palmieri G.
Role of key-regulator genes in melanoma susceptibility and pathogenesis among patients from South Italy.
BMC Cancer 9:352.1-352.11(2009)
PubMed=20041153; DOI=10.1371/journal.pone.0008461
Jeffs A.R., Glover A.C., Slobbe L.J., Wang L., He S.-J., Hazlett J.A., Awasthi A., Woolley A.G., Marshall E.S., Joseph W.R., Print C.G., Baguley B.C., Eccles M.R.
A gene expression signature of invasive potential in metastatic melanoma cells.
PLoS ONE 4:E8461-E8461(2009)
PubMed=20164919; DOI=10.1038/nature08768
Bignell G.R., Greenman C.D., Davies H., Butler A.P., Edkins S., Andrews J.M., Buck G., Chen L., Beare D., Latimer C., Widaa S., Hinton J., Fahey C., Fu B.-Y., Swamy S., Dalgliesh G.L., Teh B.T., Deloukas P., Yang F.-T., Campbell P.J., Futreal P.A., Stratton M.R.
Signatures of mutation and selection in the cancer genome.
Nature 463:893-898(2010)
PubMed=20406486; DOI=10.1186/1479-5876-8-39
Sondergaard J.N., Nazarian R., Wang Q., Guo D.-L., Hsueh T., Mok S., Sazegar H., MacConaill L.E., Barretina J.G., Kehoe S.M., Attar N., von Euw E.M., Zuckerman J.E., Chmielowski B., Comin-Anduix B., Koya R.C., Mischel P.S., Lo R.S., Ribas A.
Differential sensitivity of melanoma cell lines with BRAFV600E mutation to the specific Raf inhibitor PLX4032.
J. Transl. Med. 8:39.1-39.11(2010)
PubMed=21424129; DOI=10.3892/or.2011.1220
Manca A., Sini M.C., Izzo F., Ascierto P.A., Tatangelo F., Botti G., Gentilcore G., Capone M., Mozzillo N., Rozzo C., Cossu A., Tanda F., Palmieri G.
Induction of arginosuccinate synthetase (ASS) expression affects the antiproliferative activity of arginine deiminase (ADI) in melanoma cells.
Oncol. Rep. 25:1495-1502(2011)
PubMed=22068913; DOI=10.1073/pnas.1111840108
Gillet J.-P., Calcagno A.M., Varma S., Marino M., Green L.J., Vora M.I., Patel C., Orina J.N., Eliseeva T.A., Singal V., Padmanabhan R., Davidson B., Ganapathi R., Sood A.K., Rueda B.R., Ambudkar S.V., Gottesman M.M.
Redefining the relevance of established cancer cell lines to the study of mechanisms of clinical anti-cancer drug resistance.
Proc. Natl. Acad. Sci. U.S.A. 108:18708-18713(2011)
PubMed=21725359; DOI=10.1038/onc.2011.250
Xing F., Persaud Y., Pratilas C.A., Taylor B.S., Janakiraman M., She Q.-B., Gallardo H., Liu C., Merghoub T., Hefter B., Dolgalev I., Viale A.J., Heguy A., de Stanchina E., Cobrinik D., Bollag G., Wolchok J.D., Houghton A.N., Solit D.B.
Concurrent loss of the PTEN and RB1 tumor suppressors attenuates RAF dependence in melanomas harboring (V600E)BRAF.
Oncogene 31:446-457(2012)
PubMed=22347499; DOI=10.1371/journal.pone.0031628
Ruan X.-Y., Kocher J.-P.A., Pommier Y., Liu H.-F., Reinhold W.C.
Mass homozygotes accumulation in the NCI-60 cancer cell lines as compared to HapMap trios, and relation to fragile site location.
PLoS ONE 7:E31628-E31628(2012)
PubMed=22384151; DOI=10.1371/journal.pone.0032096
Lee J.-S., Kim Y.K., Kim H.J., Hajar S., Tan Y.L., Kang N.-Y., Ng S.H., Yoon C.N., Chang Y.-T.
Identification of cancer cell-line origins using fluorescence image-based phenomic screening.
PLoS ONE 7:E32096-E32096(2012)
PubMed=22460905; DOI=10.1038/nature11003
Barretina J.G., Caponigro G., Stransky N., Venkatesan K., Margolin A.A., Kim S., Wilson C.J., Lehar J., Kryukov G.V., Sonkin D., Reddy A., Liu M., Murray L., Berger M.F., Monahan J.E., Morais P., Meltzer J., Korejwa A., Jane-Valbuena J., Mapa F.A., Thibault J., Bric-Furlong E., Raman P., Shipway A., Engels I.H., Cheng J., Yu G.-Y.K., Yu J.-J., Aspesi P. Jr., de Silva M., Jagtap K., Jones M.D., Wang L., Hatton C., Palescandolo E., Gupta S., Mahan S., Sougnez C., Onofrio R.C., Liefeld T., MacConaill L.E., Winckler W., Reich M., Li N.-X., Mesirov J.P., Gabriel S.B., Getz G., Ardlie K., Chan V., Myer V.E., Weber B.L., Porter J., Warmuth M., Finan P., Harris J.L., Meyerson M.L., Golub T.R., Morrissey M.P., Sellers W.R., Schlegel R., Garraway L.A.
The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity.
Nature 483:603-607(2012)
PubMed=22628656; DOI=10.1126/science.1218595
Jain M., Nilsson R., Sharma S., Madhusudhan N., Kitami T., Souza A.L., Kafri R., Kirschner M.W., Clish C.B., Mootha V.K.
Metabolite profiling identifies a key role for glycine in rapid cancer cell proliferation.
Science 336:1040-1044(2012)
PubMed=23285177; DOI=10.1371/journal.pone.0052760
Schayowitz A.B., Bertenshaw G.P., Jeffries E., Schatz T., Cotton J., Villanueva J., Herlyn M., Krepler C., Vultur A., Xu W., Yu G.H., Schuchter L.M., Clark D.P.
Functional profiling of live melanoma samples using a novel automated platform.
PLoS ONE 7:E52760-E52760(2012)
PubMed=23856246; DOI=10.1158/0008-5472.CAN-12-3342
Abaan O.D., Polley E.C., Davis S.R., Zhu Y.-L.J., Bilke S., Walker R.L., Pineda M.A., Gindin Y., Jiang Y., Reinhold W.C., Holbeck S.L., Simon R.M., Doroshow J.H., Pommier Y., Meltzer P.S.
The exomes of the NCI-60 panel: a genomic resource for cancer biology and systems pharmacology.
Cancer Res. 73:4372-4382(2013)
PubMed=23933261; DOI=10.1016/j.celrep.2013.07.018
Moghaddas Gholami A., Hahne H., Wu Z.-X., Auer F.J., Meng C., Wilhelm M., Kuster B.
Global proteome analysis of the NCI-60 cell line panel.
Cell Rep. 4:609-620(2013)
PubMed=24279929; DOI=10.1186/2049-3002-1-20
Dolfi S.C., Chan L.L.-Y., Qiu J., Tedeschi P.M., Bertino J.R., Hirshfield K.M., Oltvai Z.N., Vazquez A.
The metabolic demands of cancer cells are coupled to their size and protein synthesis rates.
Cancer Metab. 1:20.1-20.13(2013)
PubMed=24576830; DOI=10.1158/0008-5472.CAN-13-2625
Nissan M.H., Pratilas C.A., Jones A.M., Ramirez R., Won H., Liu C.-L., Tiwari S., Kong L., Hanrahan A.J., Yao Z., Merghoub T., Ribas A., Chapman P.B., Yaeger R., Taylor B.S., Schultz N., Berger M.F., Rosen N., Solit D.B.
Loss of NF1 in cutaneous melanoma is associated with RAS activation and MEK dependence.
Cancer Res. 74:2340-2350(2014)
PubMed=24670534; DOI=10.1371/journal.pone.0092047
Varma S., Pommier Y., Sunshine M., Weinstein J.N., Reinhold W.C.
High resolution copy number variation data in the NCI-60 cancer cell lines from whole genome microarrays accessible through CellMiner.
PLoS ONE 9:E92047-E92047(2014)
PubMed=25960936; DOI=10.4161/21624011.2014.954893
Boegel S., Lower M., Bukur T., Sahin U., Castle J.C.
A catalog of HLA type, HLA expression, and neo-epitope candidates in human cancer cell lines.
OncoImmunology 3:e954893.1-e954893.12(2014)
PubMed=25485619; DOI=10.1038/nbt.3080
Klijn C., Durinck S., Stawiski E.W., Haverty P.M., Jiang Z.-S., Liu H.-B., Degenhardt J., Mayba O., Gnad F., Liu J.-F., Pau G., Reeder J., Cao Y., Mukhyala K., Selvaraj S.K., Yu M.-M., Zynda G.J., Brauer M.J., Wu T.D., Gentleman R.C., Manning G., Yauch R.L., Bourgon R., Stokoe D., Modrusan Z., Neve R.M., de Sauvage F.J., Settleman J., Seshagiri S., Zhang Z.-M.
A comprehensive transcriptional portrait of human cancer cell lines.
Nat. Biotechnol. 33:306-312(2015)
PubMed=25877200; DOI=10.1038/nature14397
Yu M., Selvaraj S.K., Liang-Chu M.M.Y., Aghajani S., Busse M., Yuan J., Lee G., Peale F.V., Klijn C., Bourgon R., Kaminker J.S., Neve R.M.
A resource for cell line authentication, annotation and quality control.
Nature 520:307-311(2015)
PubMed=25950383; DOI=10.1111/pcmr.12380
Lazar I., Clement E., Ducoux-Petit M., Denat L., Soldan V., Dauvillier S., Balor S., Burlet-Schiltz O., Larue L., Muller C., Nieto L.
Proteome characterization of melanoma exosomes reveals a specific signature for metastatic cell lines.
Pigment Cell Melanoma Res. 28:464-475(2015)
PubMed=26405815; DOI=10.1371/journal.pone.0138210
Capaldo B.J., Roller D.G., Axelrod M.J., Koeppel A.F., Petricoin E.F. III, Slingluff C.L. Jr., Weber M.J., Mackey A.J., Gioeli D., Bekiranov S.
Systems analysis of adaptive responses to MAP kinase pathway blockade in BRAF mutant melanoma.
PLoS ONE 10:E0138210-E0138210(2015)
PubMed=26589293; DOI=10.1186/s13073-015-0240-5
Scholtalbers J., Boegel S., Bukur T., Byl M., Goerges S., Sorn P., Loewer M., Sahin U., Castle J.C.
TCLP: an online cancer cell line catalogue integrating HLA type, predicted neo-epitopes, virus and gene expression.
Genome Med. 7:118.1-118.7(2015)
PubMed=26673621; DOI=10.18632/oncotarget.6548
Roller D.G., Capaldo B.J., Bekiranov S., Mackey A.J., Conaway M.R., Petricoin E.F. III, Gioeli D., Weber M.J.
Combinatorial drug screening and molecular profiling reveal diverse mechanisms of intrinsic and adaptive resistance to BRAF inhibition in V600E BRAF mutant melanomas.
Oncotarget 7:2734-2753(2016)
PubMed=27087056; DOI=10.1038/srep24569
Haridas P., McGovern J.A., Kashyap A.S., McElwain D.L.S., Simpson M.J.
Standard melanoma-associated markers do not identify the MM127 metastatic melanoma cell line.
Sci. Rep. 6:24569-24569(2016)
PubMed=27377824; DOI=10.1038/sdata.2016.52
Mestdagh P., Lefever S., Volders P.-J., Derveaux S., Hellemans J., Vandesompele J.
Long non-coding RNA expression profiling in the NCI60 cancer cell line panel using high-throughput RT-qPCR.
Sci. Data 3:160052-160052(2016)
PubMed=27397505; DOI=10.1016/j.cell.2016.06.017
Iorio F., Knijnenburg T.A., Vis D.J., Bignell G.R., Menden M.P., Schubert M., Aben N., Goncalves E., Barthorpe S., Lightfoot H., Cokelaer T., Greninger P., van Dyk E., Chang H., de Silva H., Heyn H., Deng X.-M., Egan R.K., Liu Q.-S., Mironenko T., Mitropoulos X., Richardson L., Wang J.-H., Zhang T.-H., Moran S., Sayols S., Soleimani M., Tamborero D., Lopez-Bigas N., Ross-Macdonald P., Esteller M., Gray N.S., Haber D.A., Stratton M.R., Benes C.H., Wessels L.F.A., Saez-Rodriguez J., McDermott U., Garnett M.J.
A landscape of pharmacogenomic interactions in cancer.
Cell 166:740-754(2016)
PubMed=27807467; DOI=10.1186/s13100-016-0078-4
Zampella J.G., Rodic N., Yang W.R., Huang C.R.L., Welch J., Gnanakkan V.P., Cornish T.C., Boeke J.D., Burns K.H.
A map of mobile DNA insertions in the NCI-60 human cancer cell panel.
Mob. DNA 7:20.1-20.11(2016)
PubMed=28196595; DOI=10.1016/j.ccell.2017.01.005
Li J., Zhao W., Akbani R., Liu W.-B., Ju Z.-L., Ling S.-Y., Vellano C.P., Roebuck P., Yu Q.-H., Eterovic A.K., Byers L.A., Davies M.A., Deng W.-L., Gopal Y.N.V., Chen G., von Euw E.M., Slamon D.J., Conklin D., Heymach J.V., Gazdar A.F., Minna J.D., Myers J.N., Lu Y.-L., Mills G.B., Liang H.
Characterization of human cancer cell lines by reverse-phase protein arrays.
Cancer Cell 31:225-239(2017)
PubMed=29492214; DOI=10.18632/oncotarget.23989
Sini M.C., Doneddu V., Paliogiannis P., Casula M., Colombino M., Manca A., Botti G., Ascierto P.A., Lissia A., Cossu A., Palmieri G.
Genetic alterations in main candidate genes during melanoma progression.
Oncotarget 9:8531-8541(2018)
PubMed=30674989; DOI=10.1038/s41388-018-0640-2
Mancuso P., Tricarico R., Bhattacharjee V., Cosentino L., Kadariya Y., Jelinek J., Nicolas E., Einarson M., Beeharry N., Devarajan K., Katz R.A., Dorjsuren D.G., Sun H.-M., Simeonov A., Giordano A., Testa J.R., Davidson G., Davidson I., Larue L., Sobol R.W., Yen T.J., Bellacosa A.
Thymine DNA glycosylase as a novel target for melanoma.
Oncogene 38:3710-3728(2019)
PubMed=30894373; DOI=10.1158/0008-5472.CAN-18-2747
Dutil J., Chen Z.-H., Monteiro A.N.A., Teer J.K., Eschrich S.A.
An interactive resource to probe genetic diversity and estimated ancestry in cancer cell lines.
Cancer Res. 79:1263-1273(2019)
PubMed=31068700; DOI=10.1038/s41586-019-1186-3
Ghandi M., Huang F.W., Jane-Valbuena J., Kryukov G.V., Lo C.C., McDonald E.R. III, Barretina J.G., Gelfand E.T., Bielski C.M., Li H.-X., Hu K., Andreev-Drakhlin A.Y., Kim J., Hess J.M., Haas B.J., Aguet F., Weir B.A., Rothberg M.V., Paolella B.R., Lawrence M.S., Akbani R., Lu Y.-L., Tiv H.L., Gokhale P.C., de Weck A., Mansour A.A., Oh C., Shih J., Hadi K., Rosen Y., Bistline J., Venkatesan K., Reddy A., Sonkin D., Liu M., Lehar J., Korn J.M., Porter D.A., Jones M.D., Golji J., Caponigro G., Taylor J.E., Dunning C.M., Creech A.L., Warren A.C., McFarland J.M., Zamanighomi M., Kauffmann A., Stransky N., Imielinski M., Maruvka Y.E., Cherniack A.D., Tsherniak A., Vazquez F., Jaffe J.D., Lane A.A., Weinstock D.M., Johannessen C.M., Morrissey M.P., Stegmeier F., Schlegel R., Hahn W.C., Getz G., Mills G.B., Boehm J.S., Golub T.R., Garraway L.A., Sellers W.R.
Next-generation characterization of the Cancer Cell Line Encyclopedia.
Nature 569:503-508(2019)
PubMed=31978347; DOI=10.1016/j.cell.2019.12.023
Nusinow D.P., Szpyt J., Ghandi M., Rose C.M., McDonald E.R. III, Kalocsay M., Jane-Valbuena J., Gelfand E.T., Schweppe D.K., Jedrychowski M.P., Golji J., Porter D.A., Rejtar T., Wang Y.K., Kryukov G.V., Stegmeier F., Erickson B.K., Garraway L.A., Sellers W.R., Gygi S.P.
Quantitative proteomics of the Cancer Cell Line Encyclopedia.
Cell 180:387-402.e16(2020)
PubMed=35839778; DOI=10.1016/j.ccell.2022.06.010
Goncalves E., Poulos R.C., Cai Z.-X., Barthorpe S., Manda S.S., Lucas N., Beck A., Bucio-Noble D., Dausmann M., Hall C., Hecker M., Koh J., Lightfoot H., Mahboob S., Mali I., Morris J., Richardson L., Seneviratne A.J., Shepherd R., Sykes E., Thomas F., Valentini S., Williams S.G., Wu Y.-X., Xavier D., MacKenzie K.L., Hains P.G., Tully B., Robinson P.J., Zhong Q., Garnett M.J., Reddel R.R.
Pan-cancer proteomic map of 949 human cell lines.
Cancer Cell 40:835-849.e8(2022)

 上海中乔新舟生物科技有限公司
上海中乔新舟生物科技有限公司