配套完培,省时省力,单买细胞无优惠
|
产品名称 |
LN229人大脑神经母瘤细胞 |
|
货号 |
ZQ1001 |
|
产品介绍 |
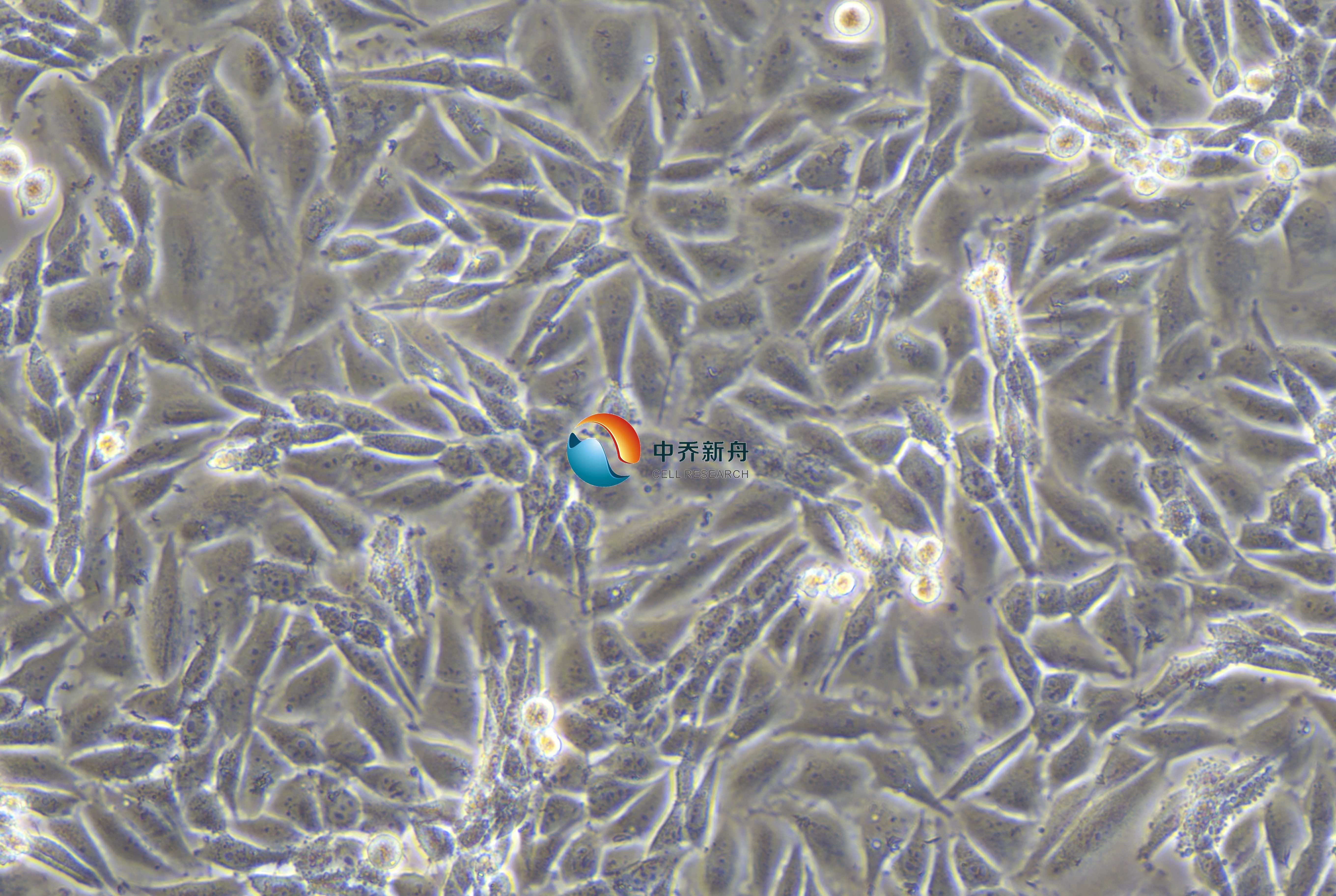
LN229细胞是一种人类胶质母细胞瘤(glioblastoma multiforme, GBM)细胞系,最初在1979年从一位60岁白人女性患者的右额顶枕胶质母细胞瘤中建立。胶质母细胞瘤是最具侵袭性和致死性的脑癌之一,LN229细胞被广泛用于研究,以了解该疾病的分子基础并制定潜在的治疗策略。细胞表现出上皮样形态和粘附生长特性,这使它成为体外研究的理想选择。考虑到它的高致瘤性,当注射到裸鼠体内时,很容易形成肿瘤,使其成为癌症研究的强大模型。
LN229细胞的关键特征之一是存在突变的p53基因(TP53),在密码子98处具有特异性的CCT (Pro)到CTT (Leu)突变。这种突变对细胞系的攻击行为和对凋亡的抵抗起着重要作用。此外,LN229细胞有一个野生型PTEN基因,但它们在p16和p14ARF肿瘤抑制基因中表现出纯合缺失,而p16和p14ARF是细胞周期和凋亡的重要调节因子。这些遗传改变使LN229成为研究这些突变对肿瘤生物学和治疗耐药性影响的有价值的模型。 |
|
种属 |
人 |
|
性别/年龄 |
女性/60岁 |
|
组织 |
脑/右额叶顶枕皮层 |
|
疾病 |
胶质母细胞瘤 |
|
生物安全等级 |
BSL-1 |
|
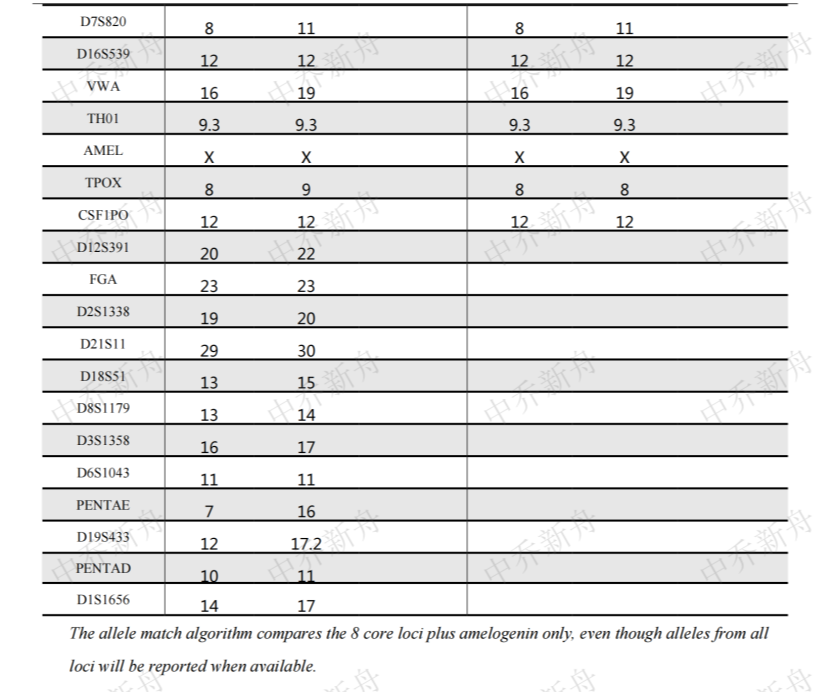
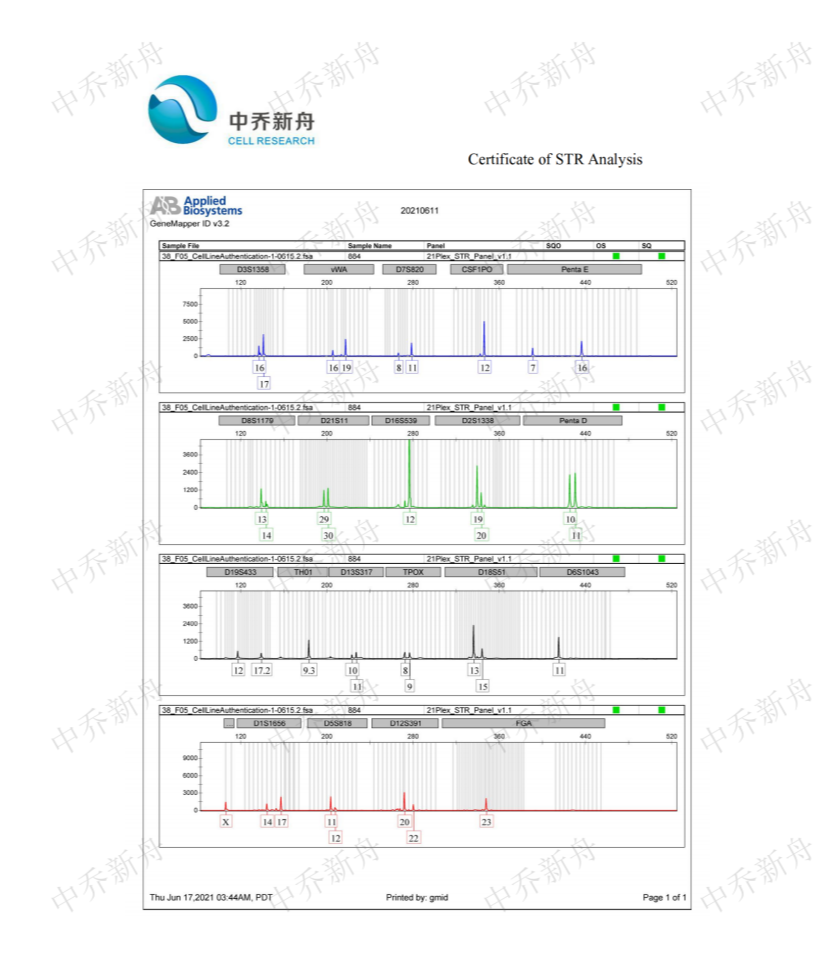
STR位点信息 |
Amelogenin:X CSF1PO:12 D13S317:10,11 D16S539:12 D5S818:11,12 D7S820:8,11 TH01:9.3 TPOX:8 vWA:16,19 |
|
细胞类型 |
胶质母细胞瘤 |
|
形态学 |
上皮样 |
|
生长方式 |
贴壁生长 |
|
倍增时间 |
|
|
培养基和添加剂 |
DMEM(中乔新舟 货号:ZQ-100)+5%FBS(中乔新舟 货号:ZQ500-A)+1%PS(中乔新舟 货号:CSP006) |
|
推荐完全培养基货号 |
|
|
培养条件 |
95%空气,5%二氧化碳;37℃ |
|
抗原表达/受体表达 |
*** |
|
基因表达 |
*** |
|
保藏机构 |
ATCC;CRL-2611 |
|
供应限制 |
仅供科研使用 |
|
货号 |
ZQ1001 |
|
发货规格 |
活细胞:T25培养瓶*1瓶或者1ml 冻存管*2支(细胞量约为1x10^6 Cells/Vial )二选一 |
|
发货形式 |
活细胞:常温运输;冻存管:干冰运输 |
|
储存温度 |
活细胞:培养箱;冻存管:液氮罐 |
|
产地 |
中国 |
|
供应限制 |
仅供科研使用 |
PubMed=7693337
Li H., Hamou M.-F., de Tribolet N., Jaufeerally R., Hofmann M., Diserens A.-C., Van Meir E.G.
Variant CD44 adhesion molecules are expressed in human brain metastases but not in glioblastomas.
Cancer Res. 53:5345-5349(1993)
PubMed=8509230; DOI=10.1002/ijc.2910540329
Rimoldi D., Romero P., Carrel S.
The human melanoma antigen-encoding gene, MAGE-1, is expressed by other tumour cells of neuroectodermal origin such as glioblastomas and neuroblastomas.
Int. J. Cancer 54:527-528(1993)
PubMed=9842975; DOI=10.1002/(SICI)1097-0215(19981218)79:6<640::AID-IJC15>3.0.CO;2-z
Weller M., Rieger J., Grimmel C., Van Meir E.G., De Tribolet N., Krajewski S., Reed J.C., von Deimling A., Dichgans J.
Predicting chemoresistance in human malignant glioma cells: the role of molecular genetic analyses.
Int. J. Cancer 79:640-644(1998)
DOI=10.1007/0-306-46861-1_11
Ali-Osman F.
Brain tumors.
(In) Human cell culture. Vol. 2. Cancer cell lines part 2; Masters J.R.W., Palsson B.O. (eds.); pp.167-184; Kluwer Academic Publishers; New York (1999)
PubMed=10416987; DOI=10.1111/j.1750-3639.1999.tb00536.x
Ishii N., Maier D., Merlo A., Tada M., Sawamura Y., Diserens A.-C., Van Meir E.G.
Frequent co-alterations of TP53, p16/CDKN2A, p14ARF, PTEN tumor suppressor genes in human glioma cell lines.
Brain Pathol. 9:469-479(1999)
PubMed=14614447; DOI=10.1038/sj.onc.1207198
Wischhusen J., Naumann U., Ohgaki H., Rastinejad F., Weller M.
CP-31398, a novel p53-stabilizing agent, induces p53-dependent and p53-independent glioma cell death.
Oncogene 22:8233-8245(2003)
PubMed=14655754; DOI=10.1111/j.1750-3639.2003.tb00479.x
Bahr O., Rieger J., Duffner F., Meyermann R., Weller M., Wick W.
P-glycoprotein and multidrug resistance-associated protein mediate specific patterns of multidrug resistance in malignant glioma cell lines, but not in primary glioma cells.
Brain Pathol. 13:482-494(2003)
PubMed=16697959; DOI=10.1016/j.ccr.2006.03.030
Lee J., Kotliarova S., Kotliarov Y., Li A.-G., Su Q., Donin N.M., Pastorino S., Purow B.W., Christopher N., Zhang W., Park J.K., Fine H.A.
Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines.
Cancer Cell 9:391-403(2006)
PubMed=17595512; DOI=10.1159/000104150
Rieger J., Frank B., Weller M., Wick W.
Mechanisms of resistance of human glioma cells to Apo2 ligand/TNF-related apoptosis-inducing ligand.
Cell. Physiol. Biochem. 20:23-34(2007)
PubMed=19365568; DOI=10.1371/journal.pone.0005209
Bax D.A., Little S.E., Gaspar N., Perryman L., Marshall L., Viana-Pereira M., Jones T.A., Williams R.D., Grigoriadis A., Vassal G., Workman P., Sheer D., Reis R.M., Pearson A.D.J., Hargrave D., Jones C.
Molecular and phenotypic characterisation of paediatric glioma cell lines as models for preclinical drug development.
PLoS ONE 4:E5209-E5209(2009)
PubMed=20504876; DOI=10.1093/neuonc/noq051
Berger B., Capper D., Lemke D., Pfenning P.-N., Platten M., Weller M., von Deimling A., Wick W., Weiler M.
Defective p53 antiangiogenic signaling in glioblastoma.
Neuro-oncol. 12:894-907(2010)
PubMed=21406405; DOI=10.1158/0008-5472.CAN-10-3112
Grzmil M., Morin P. Jr., Lino M.M., Merlo A., Frank S., Wang Y.-H., Moncayo G., Hemmings B.A.
MAP kinase-interacting kinase 1 regulates SMAD2-dependent TGF-beta signaling pathway in human glioblastoma.
Cancer Res. 71:2392-2402(2011)
PubMed=22460905; DOI=10.1038/nature11003
Barretina J.G., Caponigro G., Stransky N., Venkatesan K., Margolin A.A., Kim S., Wilson C.J., Lehar J., Kryukov G.V., Sonkin D., Reddy A., Liu M., Murray L., Berger M.F., Monahan J.E., Morais P., Meltzer J., Korejwa A., Jane-Valbuena J., Mapa F.A., Thibault J., Bric-Furlong E., Raman P., Shipway A., Engels I.H., Cheng J., Yu G.-Y.K., Yu J.-J., Aspesi P. Jr., de Silva M., Jagtap K., Jones M.D., Wang L., Hatton C., Palescandolo E., Gupta S., Mahan S., Sougnez C., Onofrio R.C., Liefeld T., MacConaill L.E., Winckler W., Reich M., Li N.-X., Mesirov J.P., Gabriel S.B., Getz G., Ardlie K., Chan V., Myer V.E., Weber B.L., Porter J., Warmuth M., Finan P., Harris J.L., Meyerson M.L., Golub T.R., Morrissey M.P., Sellers W.R., Schlegel R., Garraway L.A.
The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity.
Nature 483:603-607(2012)
PubMed=22570425; DOI=10.1093/neuonc/nos072
Bady P., Diserens A.-C., Castella V., Kalt S., Heinimann K., Hamou M.-F., Delorenzi M., Hegi M.E.
DNA fingerprinting of glioma cell lines and considerations on similarity measurements.
Neuro-oncol. 14:701-711(2012)
PubMed=25984343; DOI=10.1038/sdata.2014.35
Cowley G.S., Weir B.A., Vazquez F., Tamayo P., Scott J.A., Rusin S., East-Seletsky A., Ali L.D., Gerath W.F.J., Pantel S.E., Lizotte P.H., Jiang G.-Z., Hsiao J., Tsherniak A., Dwinell E., Aoyama S., Okamoto M., Harrington W., Gelfand E.T., Green T.M., Tomko M.J., Gopal S., Wong T.C., Li H.-B., Howell S., Stransky N., Liefeld T., Jang D., Bistline J., Meyers B.H., Armstrong S.A., Anderson K.C., Stegmaier K., Reich M., Pellman D., Boehm J.S., Mesirov J.P., Golub T.R., Root D.E., Hahn W.C.
Parallel genome-scale loss of function screens in 216 cancer cell lines for the identification of context-specific genetic dependencies.
Sci. Data 1:140035-140035(2014)
PubMed=25485619; DOI=10.1038/nbt.3080
Klijn C., Durinck S., Stawiski E.W., Haverty P.M., Jiang Z.-S., Liu H.-B., Degenhardt J., Mayba O., Gnad F., Liu J.-F., Pau G., Reeder J., Cao Y., Mukhyala K., Selvaraj S.K., Yu M.-M., Zynda G.J., Brauer M.J., Wu T.D., Gentleman R.C., Manning G., Yauch R.L., Bourgon R., Stokoe D., Modrusan Z., Neve R.M., de Sauvage F.J., Settleman J., Seshagiri S., Zhang Z.-M.
A comprehensive transcriptional portrait of human cancer cell lines.
Nat. Biotechnol. 33:306-312(2015)
PubMed=25877200; DOI=10.1038/nature14397
Yu M., Selvaraj S.K., Liang-Chu M.M.Y., Aghajani S., Busse M., Yuan J., Lee G., Peale F.V., Klijn C., Bourgon R., Kaminker J.S., Neve R.M.
A resource for cell line authentication, annotation and quality control.
Nature 520:307-311(2015)
PubMed=25894527; DOI=10.1371/journal.pone.0121314
Bausch-Fluck D., Hofmann A., Bock T., Frei A.P., Cerciello F., Jacobs A., Moest H., Omasits U., Gundry R.L., Yoon C., Schiess R., Schmidt A., Mirkowska P., Hartlova A.S., Van Eyk J.E., Bourquin J.-P., Aebersold R., Boheler K.R., Zandstra P.W., Wollscheid B.
A mass spectrometric-derived cell surface protein atlas.
PLoS ONE 10:E0121314-E0121314(2015)
PubMed=26496030; DOI=10.18632/oncotarget.6171
Patil V., Pal J., Somasundaram K.
Elucidating the cancer-specific genetic alteration spectrum of glioblastoma derived cell lines from whole exome and RNA sequencing.
Oncotarget 6:43452-43471(2015)
PubMed=26589293; DOI=10.1186/s13073-015-0240-5
Scholtalbers J., Boegel S., Bukur T., Byl M., Goerges S., Sorn P., Loewer M., Sahin U., Castle J.C.
TCLP: an online cancer cell line catalogue integrating HLA type, predicted neo-epitopes, virus and gene expression.
Genome Med. 7:118.1-118.7(2015)
PubMed=25772239; DOI=10.1038/onc.2015.61
Vassallo I., Zinn P.O., Lai M., Rajakannu P., Hamou M.-F., Hegi M.E.
WIF1 re-expression in glioblastoma inhibits migration through attenuation of non-canonical WNT signaling by downregulating the lncRNA MALAT1.
Oncogene 35:12-21(2016)
PubMed=27397505; DOI=10.1016/j.cell.2016.06.017
Iorio F., Knijnenburg T.A., Vis D.J., Bignell G.R., Menden M.P., Schubert M., Aben N., Goncalves E., Barthorpe S., Lightfoot H., Cokelaer T., Greninger P., van Dyk E., Chang H., de Silva H., Heyn H., Deng X.-M., Egan R.K., Liu Q.-S., Mironenko T., Mitropoulos X., Richardson L., Wang J.-H., Zhang T.-H., Moran S., Sayols S., Soleimani M., Tamborero D., Lopez-Bigas N., Ross-Macdonald P., Esteller M., Gray N.S., Haber D.A., Stratton M.R., Benes C.H., Wessels L.F.A., Saez-Rodriguez J., McDermott U., Garnett M.J.
A landscape of pharmacogenomic interactions in cancer.
Cell 166:740-754(2016)
PubMed=27412690; DOI=10.1074/mcp.M116.060350
Shraibman B., Kadosh D.M., Barnea E., Admon A.
Human leukocyte antigen (HLA) peptides derived from tumor antigens induced by inhibition of DNA methylation for development of drug-facilitated immunotherapy.
Mol. Cell. Proteomics 15:3058-3070(2016)
PubMed=30894373; DOI=10.1158/0008-5472.CAN-18-2747
Dutil J., Chen Z.-H., Monteiro A.N.A., Teer J.K., Eschrich S.A.
An interactive resource to probe genetic diversity and estimated ancestry in cancer cell lines.
Cancer Res. 79:1263-1273(2019)
PubMed=30971826; DOI=10.1038/s41586-019-1103-9
Behan F.M., Iorio F., Picco G., Goncalves E., Beaver C.M., Migliardi G., Santos R., Rao Y., Sassi F., Pinnelli M., Ansari R., Harper S., Jackson D.A., McRae R., Pooley R., Wilkinson P., van der Meer D.J., Dow D., Buser-Doepner C.A., Bertotti A., Trusolino L., Stronach E.A., Saez-Rodriguez J., Yusa K., Garnett M.J.
Prioritization of cancer therapeutic targets using CRISPR-Cas9 screens.
Nature 568:511-516(2019)
PubMed=31068700; DOI=10.1038/s41586-019-1186-3
Ghandi M., Huang F.W., Jane-Valbuena J., Kryukov G.V., Lo C.C., McDonald E.R. III, Barretina J.G., Gelfand E.T., Bielski C.M., Li H.-X., Hu K., Andreev-Drakhlin A.Y., Kim J., Hess J.M., Haas B.J., Aguet F., Weir B.A., Rothberg M.V., Paolella B.R., Lawrence M.S., Akbani R., Lu Y.-L., Tiv H.L., Gokhale P.C., de Weck A., Mansour A.A., Oh C., Shih J., Hadi K., Rosen Y., Bistline J., Venkatesan K., Reddy A., Sonkin D., Liu M., Lehar J., Korn J.M., Porter D.A., Jones M.D., Golji J., Caponigro G., Taylor J.E., Dunning C.M., Creech A.L., Warren A.C., McFarland J.M., Zamanighomi M., Kauffmann A., Stransky N., Imielinski M., Maruvka Y.E., Cherniack A.D., Tsherniak A., Vazquez F., Jaffe J.D., Lane A.A., Weinstock D.M., Johannessen C.M., Morrissey M.P., Stegmeier F., Schlegel R., Hahn W.C., Getz G., Mills G.B., Boehm J.S., Golub T.R., Garraway L.A., Sellers W.R.
Next-generation characterization of the Cancer Cell Line Encyclopedia.
Nature 569:503-508(2019)
PubMed=31978347; DOI=10.1016/j.cell.2019.12.023
Nusinow D.P., Szpyt J., Ghandi M., Rose C.M., McDonald E.R. III, Kalocsay M., Jane-Valbuena J., Gelfand E.T., Schweppe D.K., Jedrychowski M.P., Golji J., Porter D.A., Rejtar T., Wang Y.K., Kryukov G.V., Stegmeier F., Erickson B.K., Garraway L.A., Sellers W.R., Gygi S.P.
Quantitative proteomics of the Cancer Cell Line Encyclopedia.
Cell 180:387-402.e16(2020)
PubMed=35839778; DOI=10.1016/j.ccell.2022.06.010
Goncalves E., Poulos R.C., Cai Z.-X., Barthorpe S., Manda S.S., Lucas N., Beck A., Bucio-Noble D., Dausmann M., Hall C., Hecker M., Koh J., Lightfoot H., Mahboob S., Mali I., Morris J., Richardson L., Seneviratne A.J., Shepherd R., Sykes E., Thomas F., Valentini S., Williams S.G., Wu Y.-X., Xavier D., MacKenzie K.L., Hains P.G., Tully B., Robinson P.J., Zhong Q., Garnett M.J., Reddel R.R.
Pan-cancer proteomic map of 949 human cell lines.
Cancer Cell 40:835-849.e8(2022)

 上海中乔新舟生物科技有限公司
上海中乔新舟生物科技有限公司