|
产品名称 |
OE19人食管癌细胞 |
|
货号 |
ZQ0552 |
|
产品介绍 |
细胞系 OE19,也称为 JROECL19,于 1993 年从一名 72 岁男性患者的贲门/食管胃交界处的腺癌中建立。肿瘤被确定为病理分期 III (UICC) 并显示出中度分化。细胞系 OE19 组成型表达 HLA-A、-B 和 -C 抗原(MHC I 类)。用干扰素-γ 处理诱导 ICAM-1 (CD54) 的表达。仅在 OE19 亚群中测量了添加干扰素-γ 时 HLA-DR(MHC II 类)的表达。这些细胞表达上皮细胞角蛋白,并且在裸鼠中具有致瘤性。在 ECACC 测试时,通过短串联重复序列 (STR)-PCR 分析无法在该细胞系中检测到 Y 染色体。众所周知,由于癌细胞系的遗传不稳定性增加,Y 染色体可能会重新排列或丢失,从而导致检测不到。根据 STR-PCR 分析,细胞系与保藏者提供的来源相同。 |
|
种属 |
人 |
|
性别/年龄 |
72岁/男性 |
|
组织 |
食道/胃贲门 |
|
疾病 |
食管癌 |
|
生物安全等级 |
BSL-1 |
|
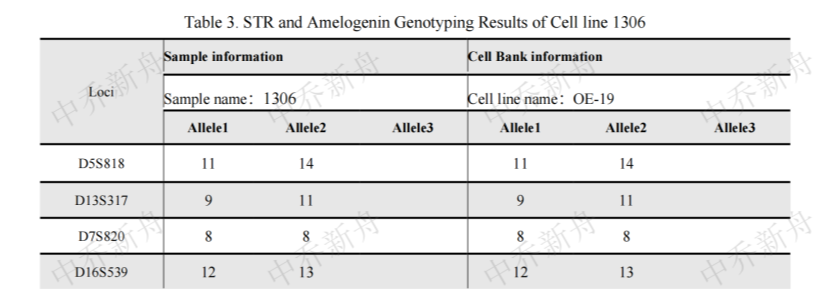
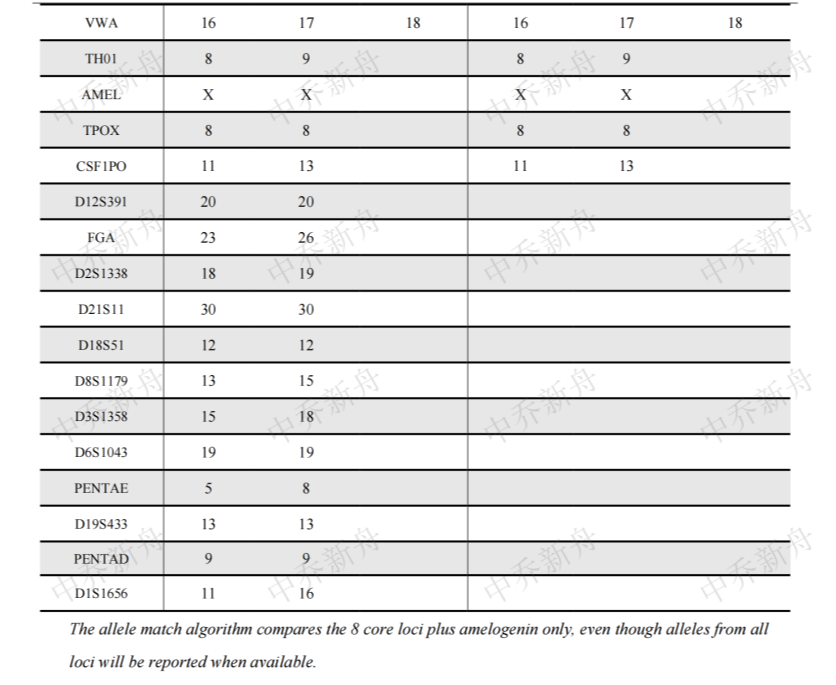
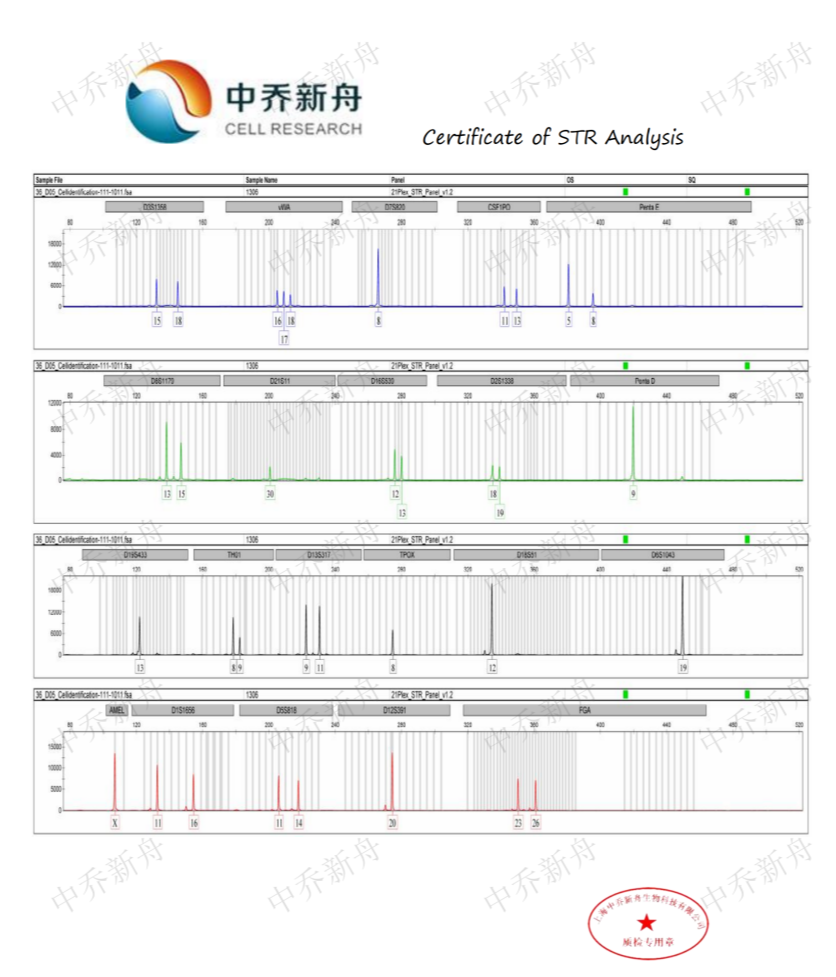
STR位点信息 |
Amelogenin: X |
|
细胞类型 |
肿瘤细胞 |
|
形态学 |
上皮样 |
|
生长方式 |
贴壁生长为单层 |
|
倍增时间 |
~50-60 hours (DSMZ=ACC-700) |
|
培养基和添加剂 |
90%RPMI-1640(中乔新舟 货号:ZQ-200)+10%胎牛血清(中乔新舟 货号:ZQ500-A)+1%双抗(品牌:中乔新舟 货号:CSP006) |
|
推荐完全培养基货号 |
|
|
基因表达 |
*** |
|
保藏机构 |
DSMZ; ACC-700ECACC; 96071721 |
|
供应限制 |
仅供科研使用 |
|
货号 |
ZQ0552 |
|
发货规格 |
活细胞:T25培养瓶*1瓶或者1ml 冻存管*2支(细胞量约为1x10^6 Cells/Vial)二选一 |
|
发货形式 |
活细胞:常温运输;冻存管:干冰运输 |
|
储存温度 |
活细胞:培养箱;冻存管:液氮罐 |
|
产地 |
中国 |
|
供应限制 |
仅供科研使用 |
PubMed=9010035; DOI=10.1038/bjc.1997.42
Rockett J.C., Larkin K., Darnton S.J., Morris A.G., Matthews H.R.
Five newly established oesophageal carcinoma cell lines: phenotypic and immunological characterization.
Br. J. Cancer 75:258-263(1997)
PubMed=15511476; DOI=10.1016/j.athoracsur.2004.05.037
Dahlberg P.S., Jacobson B.A., Dahal G., Fink J.M., Kratzke R.A., Maddaus M.A., Ferrin L.J.
ERBB2 amplifications in esophageal adenocarcinoma.
Ann. Thorac. Surg. 78:1790-1800(2004)
PubMed=16364037; DOI=10.1111/j.1442-2050.2006.00530.x
Su M., Chin S.-F., Li X.-Y., Edwards P.A.W., Caldas C., Fitzgerald R.C.
Comparative genomic hybridization of esophageal adenocarcinoma and squamous cell carcinoma cell lines.
Dis. Esophagus 19:10-14(2006)
PubMed=20075370; DOI=10.1093/jnci/djp499
Boonstra J.J., van Marion R., Beer D.G., Lin L., Chaves P., Ribeiro C., Pereira A.D., Roque L., Darnton S.J., Altorki N.K., Schrump D.S., Klimstra D.S., Tang L.H., Eshleman J.R., Alvarez H., Shimada Y., van Dekken H., Tilanus H.W., Dinjens W.N.M.
Verification and unmasking of widely used human esophageal adenocarcinoma cell lines.
J. Natl. Cancer Inst. 102:271-274(2010)
PubMed=20164919; DOI=10.1038/nature08768
Bignell G.R., Greenman C.D., Davies H., Butler A.P., Edkins S., Andrews J.M., Buck G., Chen L., Beare D., Latimer C., Widaa S., Hinton J., Fahey C., Fu B.-Y., Swamy S., Dalgliesh G.L., Teh B.T., Deloukas P., Yang F.-T., Campbell P.J., Futreal P.A., Stratton M.R.
Signatures of mutation and selection in the cancer genome.
Nature 463:893-898(2010)
PubMed=20215515; DOI=10.1158/0008-5472.CAN-09-3458
Rothenberg S.M., Mohapatra G., Rivera M.N., Winokur D., Greninger P., Nitta M., Sadow P.M., Sooriyakumar G., Brannigan B.W., Ulman M.J., Perera R.M., Wang R., Tam A., Ma X.-J., Erlander M., Sgroi D.C., Rocco J.W., Lingen M.W., Cohen E.E.W., Louis D.N., Settleman J., Haber D.A.
A genome-wide screen for microdeletions reveals disruption of polarity complex genes in diverse human cancers.
Cancer Res. 70:2158-2164(2010)
PubMed=22460905; DOI=10.1038/nature11003
Barretina J.G., Caponigro G., Stransky N., Venkatesan K., Margolin A.A., Kim S., Wilson C.J., Lehar J., Kryukov G.V., Sonkin D., Reddy A., Liu M., Murray L., Berger M.F., Monahan J.E., Morais P., Meltzer J., Korejwa A., Jane-Valbuena J., Mapa F.A., Thibault J., Bric-Furlong E., Raman P., Shipway A., Engels I.H., Cheng J., Yu G.-Y.K., Yu J.-J., Aspesi P. Jr., de Silva M., Jagtap K., Jones M.D., Wang L., Hatton C., Palescandolo E., Gupta S., Mahan S., Sougnez C., Onofrio R.C., Liefeld T., MacConaill L.E., Winckler W., Reich M., Li N.-X., Mesirov J.P., Gabriel S.B., Getz G., Ardlie K., Chan V., Myer V.E., Weber B.L., Porter J., Warmuth M., Finan P., Harris J.L., Meyerson M.L., Golub T.R., Morrissey M.P., Sellers W.R., Schlegel R., Garraway L.A.
The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity.
Nature 483:603-607(2012)
PubMed=24083764; DOI=10.7314/APJCP.2013.14.8.4891
Li J.-C., Liu D., Yang Y., Wang X.-Y., Pan D.-L., Qiu Z.-D., Su Y., Pan J.-J.
Growth, clonability, and radiation resistance of esophageal carcinoma-derived stem-like cells.
Asian Pac. J. Cancer Prev. 14:4891-4896(2013)
PubMed=25070024; DOI=10.1007/s00423-014-1235-1
Drenckhan A., Grob T., Dupree A., Dohrmann T., Mann O., Izbicki J.R., Gros S.J.
Esophageal carcinoma cell line with high EGFR polysomy is responsive to gefitinib.
Langenbecks Arch. Surg. 399:879-888(2014)
PubMed=23795680; DOI=10.1111/dote.12095
Boonstra J.J., Tilanus H.W., Dinjens W.N.M.
Translational research on esophageal adenocarcinoma: from cell line to clinic.
Dis. Esophagus 28:90-96(2015)
PubMed=25877200; DOI=10.1038/nature14397
Yu M., Selvaraj S.K., Liang-Chu M.M.Y., Aghajani S., Busse M., Yuan J., Lee G., Peale F.V., Klijn C., Bourgon R., Kaminker J.S., Neve R.M.
A resource for cell line authentication, annotation and quality control.
Nature 520:307-311(2015)
PubMed=26512696; DOI=10.3390/cancers7040881
O'Callaghan C., Fanning L.J., Barry O.P.
Hypermethylation of MAPK13 promoter in oesophageal squamous cell carcinoma is associated with loss of p38delta MAPK expression.
Cancers (Basel) 7:2124-2133(2015)
PubMed=26589293; DOI=10.1186/s13073-015-0240-5
Scholtalbers J., Boegel S., Bukur T., Byl M., Goerges S., Sorn P., Loewer M., Sahin U., Castle J.C.
TCLP: an online cancer cell line catalogue integrating HLA type, predicted neo-epitopes, virus and gene expression.
Genome Med. 7:118.1-118.7(2015)
PubMed=27397505; DOI=10.1016/j.cell.2016.06.017
Iorio F., Knijnenburg T.A., Vis D.J., Bignell G.R., Menden M.P., Schubert M., Aben N., Goncalves E., Barthorpe S., Lightfoot H., Cokelaer T., Greninger P., van Dyk E., Chang H., de Silva H., Heyn H., Deng X.-M., Egan R.K., Liu Q.-S., Mironenko T., Mitropoulos X., Richardson L., Wang J.-H., Zhang T.-H., Moran S., Sayols S., Soleimani M., Tamborero D., Lopez-Bigas N., Ross-Macdonald P., Esteller M., Gray N.S., Haber D.A., Stratton M.R., Benes C.H., Wessels L.F.A., Saez-Rodriguez J., McDermott U., Garnett M.J.
A landscape of pharmacogenomic interactions in cancer.
Cell 166:740-754(2016)
PubMed=28196595; DOI=10.1016/j.ccell.2017.01.005
Li J., Zhao W., Akbani R., Liu W.-B., Ju Z.-L., Ling S.-Y., Vellano C.P., Roebuck P., Yu Q.-H., Eterovic A.K., Byers L.A., Davies M.A., Deng W.-L., Gopal Y.N.V., Chen G., von Euw E.M., Slamon D.J., Conklin D., Heymach J.V., Gazdar A.F., Minna J.D., Myers J.N., Lu Y.-L., Mills G.B., Liang H.
Characterization of human cancer cell lines by reverse-phase protein arrays.
Cancer Cell 31:225-239(2017)
PubMed=30894373; DOI=10.1158/0008-5472.CAN-18-2747
Dutil J., Chen Z.-H., Monteiro A.N.A., Teer J.K., Eschrich S.A.
An interactive resource to probe genetic diversity and estimated ancestry in cancer cell lines.
Cancer Res. 79:1263-1273(2019)
PubMed=31068700; DOI=10.1038/s41586-019-1186-3
Ghandi M., Huang F.W., Jane-Valbuena J., Kryukov G.V., Lo C.C., McDonald E.R. III, Barretina J.G., Gelfand E.T., Bielski C.M., Li H.-X., Hu K., Andreev-Drakhlin A.Y., Kim J., Hess J.M., Haas B.J., Aguet F., Weir B.A., Rothberg M.V., Paolella B.R., Lawrence M.S., Akbani R., Lu Y.-L., Tiv H.L., Gokhale P.C., de Weck A., Mansour A.A., Oh C., Shih J., Hadi K., Rosen Y., Bistline J., Venkatesan K., Reddy A., Sonkin D., Liu M., Lehar J., Korn J.M., Porter D.A., Jones M.D., Golji J., Caponigro G., Taylor J.E., Dunning C.M., Creech A.L., Warren A.C., McFarland J.M., Zamanighomi M., Kauffmann A., Stransky N., Imielinski M., Maruvka Y.E., Cherniack A.D., Tsherniak A., Vazquez F., Jaffe J.D., Lane A.A., Weinstock D.M., Johannessen C.M., Morrissey M.P., Stegmeier F., Schlegel R., Hahn W.C., Getz G., Mills G.B., Boehm J.S., Golub T.R., Garraway L.A., Sellers W.R.
Next-generation characterization of the Cancer Cell Line Encyclopedia.
Nature 569:503-508(2019)
PubMed=31395879; DOI=10.1038/s41467-019-11415-2
Yu K., Chen B., Aran D., Charalel J., Yau C., Wolf D.M., van 't Veer L.J., Butte A.J., Goldstein T., Sirota M.
Comprehensive transcriptomic analysis of cell lines as models of primary tumors across 22 tumor types.
Nat. Commun. 10:3574.1-3574.11(2019)
PubMed=35839778; DOI=10.1016/j.ccell.2022.06.010
Goncalves E., Poulos R.C., Cai Z.-X., Barthorpe S., Manda S.S., Lucas N., Beck A., Bucio-Noble D., Dausmann M., Hall C., Hecker M., Koh J., Lightfoot H., Mahboob S., Mali I., Morris J., Richardson L., Seneviratne A.J., Shepherd R., Sykes E., Thomas F., Valentini S., Williams S.G., Wu Y.-X., Xavier D., MacKenzie K.L., Hains P.G., Tully B., Robinson P.J., Zhong Q., Garnett M.J., Reddel R.R.
Pan-cancer proteomic map of 949 human cell lines.
Cancer Cell 40:835-849.e8(2022)
 上海中乔新舟生物科技有限公司
上海中乔新舟生物科技有限公司