|
产品名称 |
NCI-H3122人非小细胞肺癌细胞 |
|
货号 |
ZQ0754 |
|
产品介绍 |
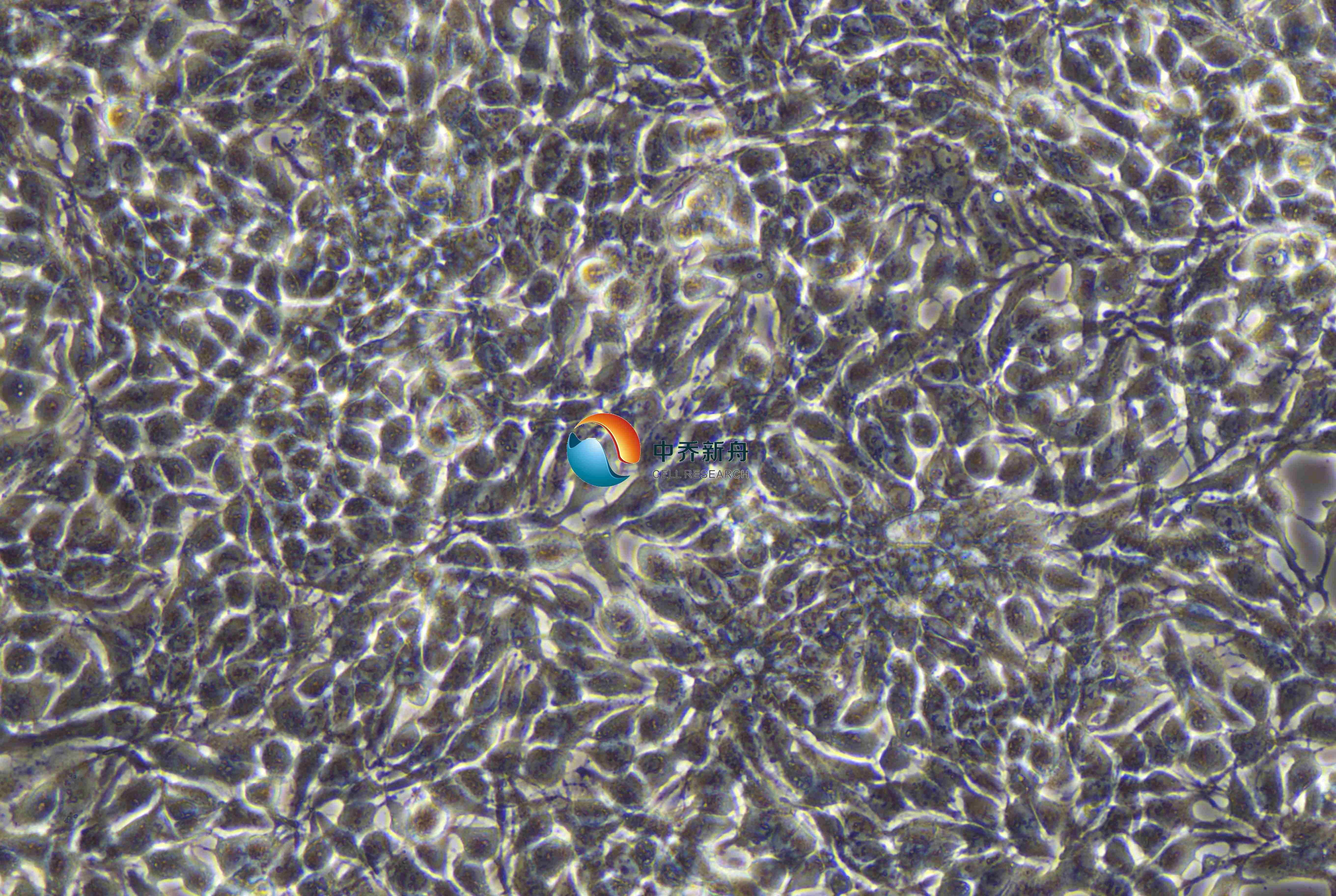
NCI-H3122细胞是一种来源于一名54岁白人男性的肺腺癌组织非小细胞肺癌(NSCLC)细胞株,由Modi等人在临床组织中分离纯化。NCI-H3122细胞的倍增时间约为48.5小时,呈现贴壁生长特性。它表达EML4-ALK融合基因,这使得它成为研究非小细胞肺癌治疗新靶点EML4-ALK的合适材料。
NCI-H3122细胞在长期接触克唑替尼后会逐渐产生耐药性,其IC50值显著提高,显示出对药物的耐受性增加。该细胞系在分子水平上表现出与非小细胞肺癌相关的基因表达特征。具体的基因突变和表达谱可能因研究而异,但通常涉及EGFR、ALK等重要的肿瘤相关基因。在药物处理后,NCI-H3122细胞的细胞周期可能会受到影响,表现出不同的增殖和凋亡特征。 |
|
种属 |
人 |
|
性别/年龄 |
男/54岁 |
|
组织 |
肺 |
|
疾病 |
非小细胞肺癌 |
|
细胞类型 |
肿瘤细胞 |
|
形态学 |
上皮样 |
|
生长方式 |
贴壁 |
|
倍增时间 |
大约48.5 hours (PubMed=29681454) |
|
培养基和添加剂 |
RPMI-1640(品牌:中乔新舟 货号:ZQ-200)+10%胎牛血清(中乔新舟 货号:ZQ500-A)+1%P/S(中乔新舟 货号:CSP006) |
|
推荐完全培养基货号 |
|
|
生物安全等级 |
BSL-1 |
|
培养条件 |
95%空气,5%二氧化碳;37℃ |
|
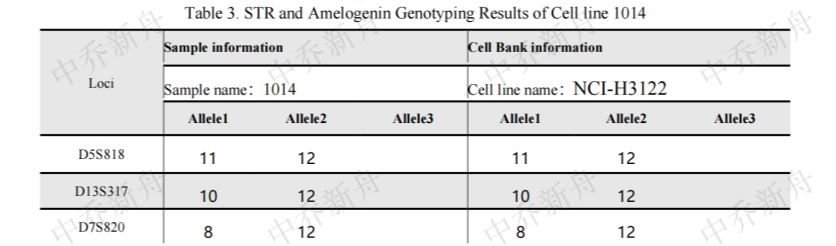
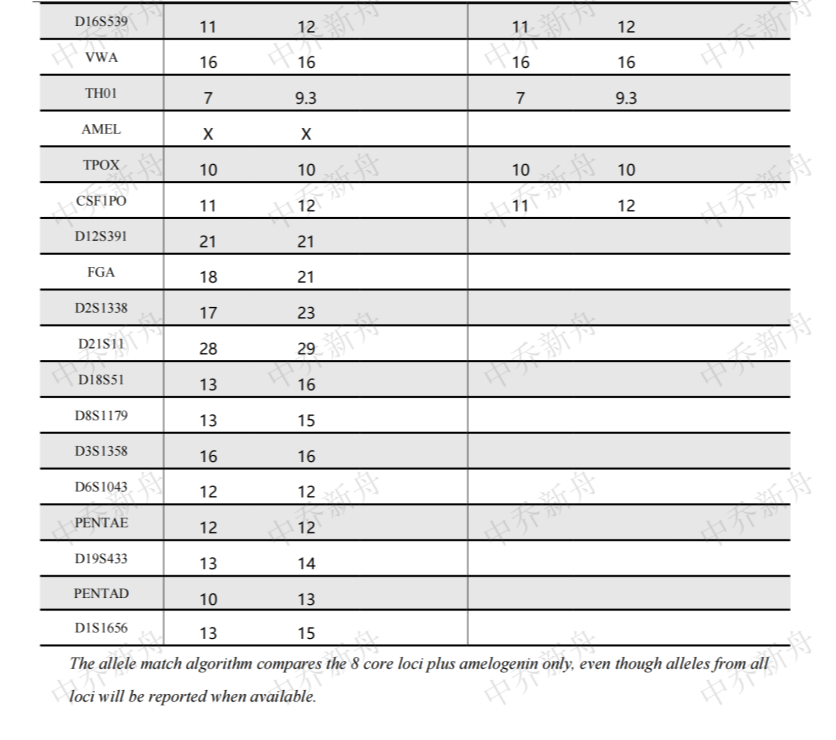
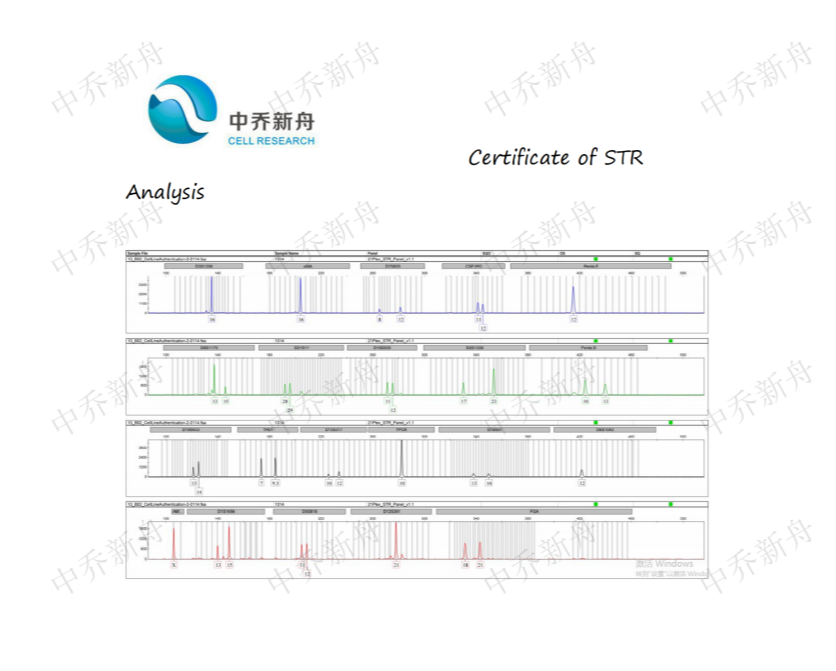
STR位点信息 |
Amelogenin:X CSF1PO: 11,12 D5S818: 11,12 D7S820: 8,12 D8S1179:13,15 D13S317:10,12 D16S539:11,12 D18S51: 13,16 FGA:18,21 Penta D: 10,13 Penta E: 12 TH01:7,9.3 TPOX:10 vWA:16 |
|
抗原表达/受体表达 |
*** |
|
基因表达 |
*** |
|
保藏机构 |
CLS; 300484 NCI-DTP; NCI-H3122 |
|
供应限制 |
仅供科研使用 |
|
货号 |
ZQ0754 |
|
发货规格 |
活细胞:T25培养瓶*1瓶或者1ml 冻存管*2支(细胞量约为1x10^6 Cells/Vial)二选一 |
|
发货形式 |
活细胞:常温运输;冻存管:干冰运输 |
|
储存温度 |
活细胞:培养箱;冻存管:液氮罐 |
|
产地 |
中国 |
|
供应限制 |
仅供科研使用 |
PubMed=11030152; DOI=10.1038/sj.onc.1203815
Modi S., Kubo A., Oie H.K., Coxon A.B., Rehmatulla A., Kaye F.J.
Protein expression of the RB-related gene family and SV40 large T antigen in mesothelioma and lung cancer.
Oncogene 19:4632-4639(2000)
PubMed=12759538; DOI=10.1159/000070299
Fujishita T., Loda M., Turner R.E., Gentler M., Kashii T., Breathnach O.S., Johnson B.E.
Sensitivity of non-small-cell lung cancer cell lines established from patients treated with prolonged infusions of paclitaxel.
Oncology 64:399-406(2003)
PubMed=18594010; DOI=10.1158/1078-0432.CCR-08-0168
Koivunen J.P., Mermel C.H., Zejnullahu K., Murphy C., Lifshits E., Holmes A.J., Choi H.G., Kim J., Chiang D., Thomas R., Lee J., Richards W.G., Sugarbaker D.J., Ducko C.T., Lindeman N.I., Marcoux J.P., Engelman J.A., Gray N.S., Lee C., Meyerson M.L., Janne P.A.
EML4-ALK fusion gene and efficacy of an ALK kinase inhibitor in lung cancer.
Clin. Cancer Res. 14:4275-4283(2008)
PubMed=20679594; DOI=10.1093/jnci/djq279
Gazdar A.F., Girard L., Lockwood W.W., Lam W.L., Minna J.D.
Lung cancer cell lines as tools for biomedical discovery and research.
J. Natl. Cancer Inst. 102:1310-1321(2010)
PubMed=22961666; DOI=10.1158/2159-8290.CD-12-0112
Byers L.A., Wang J., Nilsson M.B., Fujimoto J., Saintigny P., Yordy J., Giri U., Peyton M., Fan Y.-H., Diao L.-X., Masrorpour F., Shen L., Liu W.-B., Duchemann B., Tumula P., Bhardwaj V., Welsh J., Weber S., Glisson B.S., Kalhor N., Wistuba I.I., Girard L., Lippman S.M., Mills G.B., Coombes K.R., Weinstein J.N., Minna J.D., Heymach J.V.
Proteomic profiling identifies dysregulated pathways in small cell lung cancer and novel therapeutic targets including PARP1.
Cancer Discov. 2:798-811(2012)
PubMed=23344087; DOI=10.1097/JTO.0b013e318283dcc0
Kim S., Kim T.M., Kim D.-W., Go H., Keam B., Lee S.-H., Ku J.-L., Chung D.H., Heo D.S.
Heterogeneity of genetic changes associated with acquired crizotinib resistance in ALK-rearranged lung cancer.
J. Thorac. Oncol. 8:415-422(2013)
PubMed=24675041; DOI=10.1158/2159-8290.CD-13-0846
Friboulet L., Li N.-X., Katayama R., Lee C.C., Gainor J.F., Crystal A.S., Michellys P.-Y., Awad M.M., Yanagitani N., Kim S., Pferdekamper A.C., Li J., Kasibhatla S., Sun F., Sun X.-Y., Hua S., McNamara P., Mahmood S., Lockerman E.L., Fujita N., Nishio M., Harris J.L., Shaw A.T., Engelman J.A.
The ALK inhibitor ceritinib overcomes crizotinib resistance in non-small cell lung cancer.
Cancer Discov. 4:662-673(2014)
PubMed=26361996; DOI=10.1016/j.jprot.2015.09.003
Grundner-Culemann K., Dybowski J.N., Klammer M., Tebbe A., Schaab C., Daub H.
Comparative proteome analysis across non-small cell lung cancer cell lines.
J. Proteomics 130:1-10(2016)
PubMed=27397505; DOI=10.1016/j.cell.2016.06.017
Iorio F., Knijnenburg T.A., Vis D.J., Bignell G.R., Menden M.P., Schubert M., Aben N., Goncalves E., Barthorpe S., Lightfoot H., Cokelaer T., Greninger P., van Dyk E., Chang H., de Silva H., Heyn H., Deng X.-M., Egan R.K., Liu Q.-S., Mironenko T., Mitropoulos X., Richardson L., Wang J.-H., Zhang T.-H., Moran S., Sayols S., Soleimani M., Tamborero D., Lopez-Bigas N., Ross-Macdonald P., Esteller M., Gray N.S., Haber D.A., Stratton M.R., Benes C.H., Wessels L.F.A., Saez-Rodriguez J., McDermott U., Garnett M.J.
A landscape of pharmacogenomic interactions in cancer.
Cell 166:740-754(2016)
PubMed=29444439; DOI=10.1016/j.celrep.2018.01.051
Yuan T.L., Amzallag A., Bagni R., Yi M., Afghani S., Burgan W., Fer N., Strathern L.A., Powell K., Smith B., Waters A.M., Drubin D.A., Thomson T., Liao R., Greninger P., Stein G.T., Murchie E., Cortez E., Egan R.K., Procter L., Bess M., Cheng K.T., Lee C.-S., Lee L.C., Fellmann C., Stephens R., Luo J., Lowe S.W., Benes C.H., McCormick F.
Differential effector engagement by oncogenic KRAS.
Cell Rep. 22:1889-1902(2018)
PubMed=29681454; DOI=10.1016/j.cell.2018.03.028
McMillan E.A., Ryu M.-J., Diep C.H., Mendiratta S., Clemenceau J.R., Vaden R.M., Kim J.-H., Motoyaji T., Covington K.R., Peyton M., Huffman K., Wu X.-F., Girard L., Sung Y., Chen P.-H., Mallipeddi P.L., Lee J.Y., Hanson J., Voruganti S., Yu Y., Park S., Sudderth J., DeSevo C., Muzny D.M., Doddapaneni H., Gazdar A.F., Gibbs R.A., Hwang T.H., Heymach J.V., Wistuba I.I., Coombes K.R., Williams N.S., Wheeler D.A., MacMillan J.B., DeBerardinis R.J., Roth M.G., Posner B.A., Minna J.D., Kim H.S., White M.A.
Chemistry-first approach for nomination of personalized treatment in lung cancer.
Cell 173:864-878.e29(2018)
PubMed=30894373; DOI=10.1158/0008-5472.CAN-18-2747
Dutil J., Chen Z.-H., Monteiro A.N.A., Teer J.K., Eschrich S.A.
An interactive resource to probe genetic diversity and estimated ancestry in cancer cell lines.
Cancer Res. 79:1263-1273(2019)
PubMed=30971826; DOI=10.1038/s41586-019-1103-9
Behan F.M., Iorio F., Picco G., Goncalves E., Beaver C.M., Migliardi G., Santos R., Rao Y., Sassi F., Pinnelli M., Ansari R., Harper S., Jackson D.A., McRae R., Pooley R., Wilkinson P., van der Meer D.J., Dow D., Buser-Doepner C.A., Bertotti A., Trusolino L., Stronach E.A., Saez-Rodriguez J., Yusa K., Garnett M.J.
Prioritization of cancer therapeutic targets using CRISPR-Cas9 screens.
Nature 568:511-516(2019)
PubMed=31068700; DOI=10.1038/s41586-019-1186-3
Ghandi M., Huang F.W., Jane-Valbuena J., Kryukov G.V., Lo C.C., McDonald E.R. III, Barretina J.G., Gelfand E.T., Bielski C.M., Li H.-X., Hu K., Andreev-Drakhlin A.Y., Kim J., Hess J.M., Haas B.J., Aguet F., Weir B.A., Rothberg M.V., Paolella B.R., Lawrence M.S., Akbani R., Lu Y.-L., Tiv H.L., Gokhale P.C., de Weck A., Mansour A.A., Oh C., Shih J., Hadi K., Rosen Y., Bistline J., Venkatesan K., Reddy A., Sonkin D., Liu M., Lehar J., Korn J.M., Porter D.A., Jones M.D., Golji J., Caponigro G., Taylor J.E., Dunning C.M., Creech A.L., Warren A.C., McFarland J.M., Zamanighomi M., Kauffmann A., Stransky N., Imielinski M., Maruvka Y.E., Cherniack A.D., Tsherniak A., Vazquez F., Jaffe J.D., Lane A.A., Weinstock D.M., Johannessen C.M., Morrissey M.P., Stegmeier F., Schlegel R., Hahn W.C., Getz G., Mills G.B., Boehm J.S., Golub T.R., Garraway L.A., Sellers W.R.
Next-generation characterization of the Cancer Cell Line Encyclopedia.
Nature 569:503-508(2019)
PubMed=31803961; DOI=10.1002/jcb.29564
Mulshine J.L., Ujhazy P., Antman M., Burgess C.M., Kuzmin I.A., Bunn P.A. Jr., Johnson B.E., Roth J.A., Pass H.I., Ross S.M., Aldige C.R., Wistuba I.I., Minna J.D.
From clinical specimens to human cancer preclinical models -- a journey the NCI-cell line database-25 years later.
J. Cell. Biochem. 121:3986-3999(2020)
DOI=10.1101/2022.06.08.495301
Licciardello M.P., Zhang C., Le A.T., Doebele R.C., Clarke P.A., Workman P.
Loss of spindly sensitizes EML4-ALK v3 lung cancer cells to HSP90 inhibitors.
bioRxiv 2022:06.08.495301-06.08.495301(2022)
PubMed=35839778; DOI=10.1016/j.ccell.2022.06.010
Goncalves E., Poulos R.C., Cai Z.-X., Barthorpe S., Manda S.S., Lucas N., Beck A., Bucio-Noble D., Dausmann M., Hall C., Hecker M., Koh J., Lightfoot H., Mahboob S., Mali I., Morris J., Richardson L., Seneviratne A.J., Shepherd R., Sykes E., Thomas F., Valentini S., Williams S.G., Wu Y.-X., Xavier D., MacKenzie K.L., Hains P.G., Tully B., Robinson P.J., Zhong Q., Garnett M.J., Reddel R.R.
Pan-cancer proteomic map of 949 human cell lines.
Cancer Cell 40:835-849.e8(2022)
 上海中乔新舟生物科技有限公司
上海中乔新舟生物科技有限公司