|
产品名称 |
NCI-H660人神经内分泌前列腺癌细胞 |
|
货号 |
ZQ0838 |
|
产品介绍 |
NCI-H660是一种上皮神经内分泌细胞,从一名63岁的白人癌症男性患者的前列腺中分离出来。该细胞系由AF Gazdar和JD Minna建立,被广泛用于癌症研究,包括药物筛选、基因表达研究、细胞信号传导研究等。此外,该细胞系还可用于3D细胞培养研究,以及研究前列腺癌(特别是神经内分泌型)、小细胞癌、癌症生物学、药物筛选和开发、转移机制研究。 |
|
种属 |
人 |
|
性别/年龄 |
男/63岁 |
|
组织 |
前列腺 |
|
疾病 |
癌;小细胞肺癌;阶段E |
|
细胞类型 |
上皮神经内分泌细胞 |
|
形态学 |
上皮 |
|
生长方式 |
混合;悬浮和部分贴壁细胞 |
|
倍增时间 |
大约100 hours (PubMed=25984343) |
|
培养基和添加剂 |
RPMI-1640(ATCC改良)(中乔新舟 货号:ZQ-200F)+5%胎牛血清(中乔新舟 货号:ZQ0500)+10 nM beta-estradiol+10 nM氢化可的松(中乔新舟 货号:CSP041)+1%ITS-G(中乔新舟 货号:CSP082)+extra 2mM L-glutamine(中乔新舟 货号:CSP012)+1%P/S(中乔新舟 货号:CSP006) |
|
推荐完全培养基货号 |
|
|
生物安全等级 |
BSL-1 |
|
培养条件 |
95%空气,5%二氧化碳;37℃ |
|
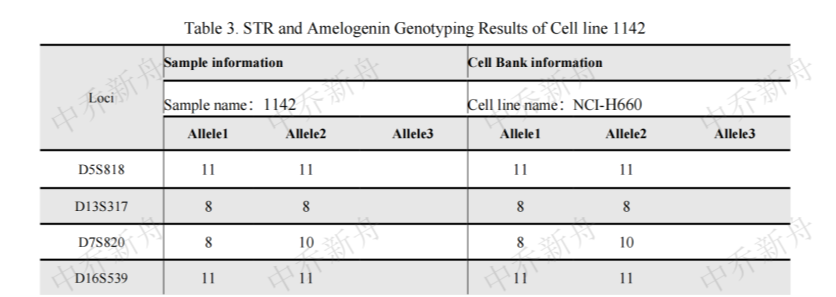
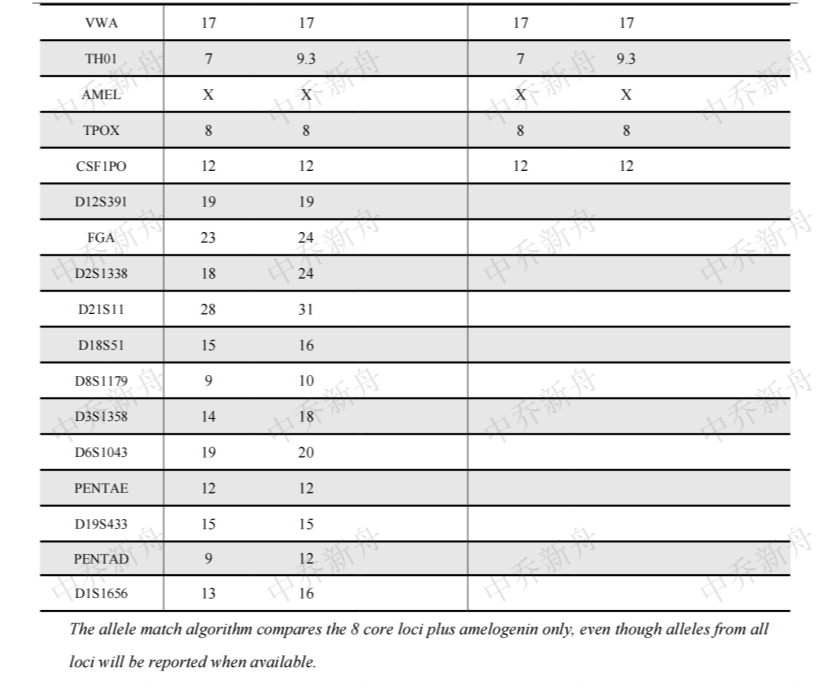
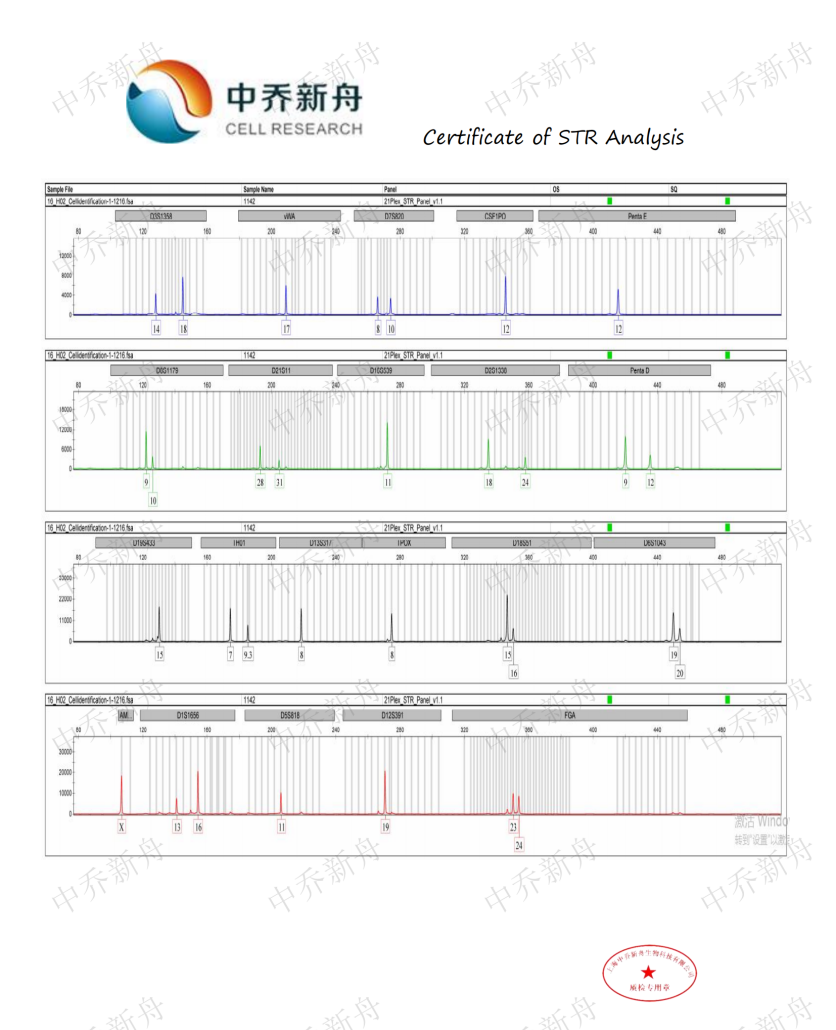
STR位点信息 |
Amelogenin: X
D3S1358: 14,18
|
|
抗原表达/受体表达 |
*** |
|
基因表达 |
*** |
|
保藏机构 |
ATCC; CRL-5813 |
|
供应限制 |
仅供科研使用 |
|
货号 |
ZQ0838 |
|
发货规格 |
活细胞:T25培养瓶*1瓶或者1ml 冻存管*2支(细胞量约为1x10^6 Cells/Vial)二选一 |
|
发货形式 |
活细胞:常温运输;冻存管:干冰运输 |
|
储存温度 |
活细胞:培养箱;冻存管:液氮罐 |
|
产地 |
中国 |
|
供应限制 |
仅供科研使用 |
PubMed=2985257
Carney D.N., Gazdar A.F., Bepler G., Guccion J.G., Marangos P.J., Moody T.W., Zweig M.H., Minna J.D.
Establishment and identification of small cell lung cancer cell lines having classic and variant features.
Cancer Res. 45:2913-2923(1985)
PubMed=2569043; DOI=10.1093/jnci/81.16.1223
Johnson B.E., Whang-Peng J., Naylor S.L., Zbar B., Brauch H., Lee E., Simmons A.M., Russell E., Nam M.H., Gazdar A.F.
Retention of chromosome 3 in extrapulmonary small cell cancer shown by molecular and cytogenetic studies.
J. Natl. Cancer Inst. 81:1223-1228(1989)
PubMed=1563005
Giaccone G., Battey J., Gazdar A.F., Oie H.K., Draoui M., Moody T.W.
Neuromedin B is present in lung cancer cell lines.
Cancer Res. 52:2732s-2736s(1992)
PubMed=8806092; DOI=10.1002/jcb.240630505
Phelps R.M., Johnson B.E., Ihde D.C., Gazdar A.F., Carbone D.P., McClintock P.R., Linnoila R.I., Matthews M.J., Bunn P.A. Jr., Carney D.N., Minna J.D., Mulshine J.L.
NCI-Navy Medical Oncology Branch cell line data base.
J. Cell. Biochem. 63 Suppl. 24:32-91(1996)
PubMed=8806103; DOI=10.1002/jcb.240630516
Johnson B.E., Russell E., Simmons A.M., Phelps R.M., Steinberg S.M., Ihde D.C., Gazdar A.F.
MYC family DNA amplification in 126 tumor cell lines from patients with small cell lung cancer.
J. Cell. Biochem. 63 Suppl. 24:210-217(1996)
PubMed=11030152; DOI=10.1038/sj.onc.1203815
Modi S., Kubo A., Oie H.K., Coxon A.B., Rehmatulla A., Kaye F.J.
Protein expression of the RB-related gene family and SV40 large T antigen in mesothelioma and lung cancer.
Oncogene 19:4632-4639(2000)
PubMed=11304728; DOI=10.1002/pros.1045
van Bokhoven A., Varella-Garcia M., Korch C.T., Hessels D., Miller G.J.
Widely used prostate carcinoma cell lines share common origins.
Prostate 47:36-51(2001)
PubMed=14518029; DOI=10.1002/pros.10290
van Bokhoven A., Varella-Garcia M., Korch C.T., Johannes W.U., Smith E.E., Miller H.L., Nordeen S.K., Miller G.J., Lucia M.S.
Molecular characterization of human prostate carcinoma cell lines.
Prostate 57:205-225(2003)
PubMed=15643172; DOI=10.1097/01.ju.0000141580.30910.57
Sobel R.E., Sadar M.D.
Cell lines used in prostate cancer research: a compendium of old and new lines -- part 1.
J. Urol. 173:342-359(2005)
PubMed=17401460; DOI=10.1593/neo.07103
Mertz K.D., Setlur S.R., Dhanasekaran S.M., Demichelis F., Perner S., Tomlins S., Tchinda J., Laxman B., Vessella R.L., Beroukhim R., Lee C., Chinnaiyan A.M., Rubin M.A.
Molecular characterization of TMPRSS2-ERG gene fusion in the NCI-H660 prostate cancer cell line: a new perspective for an old model.
Neoplasia 9:200-206(2007)
PubMed=20215515; DOI=10.1158/0008-5472.CAN-09-3458
Rothenberg S.M., Mohapatra G., Rivera M.N., Winokur D., Greninger P., Nitta M., Sadow P.M., Sooriyakumar G., Brannigan B.W., Ulman M.J., Perera R.M., Wang R., Tam A., Ma X.-J., Erlander M., Sgroi D.C., Rocco J.W., Lingen M.W., Cohen E.E.W., Louis D.N., Settleman J., Haber D.A.
A genome-wide screen for microdeletions reveals disruption of polarity complex genes in diverse human cancers.
Cancer Res. 70:2158-2164(2010)
PubMed=22460905; DOI=10.1038/nature11003
Barretina J.G., Caponigro G., Stransky N., Venkatesan K., Margolin A.A., Kim S., Wilson C.J., Lehar J., Kryukov G.V., Sonkin D., Reddy A., Liu M., Murray L., Berger M.F., Monahan J.E., Morais P., Meltzer J., Korejwa A., Jane-Valbuena J., Mapa F.A., Thibault J., Bric-Furlong E., Raman P., Shipway A., Engels I.H., Cheng J., Yu G.-Y.K., Yu J.-J., Aspesi P. Jr., de Silva M., Jagtap K., Jones M.D., Wang L., Hatton C., Palescandolo E., Gupta S., Mahan S., Sougnez C., Onofrio R.C., Liefeld T., MacConaill L.E., Winckler W., Reich M., Li N.-X., Mesirov J.P., Gabriel S.B., Getz G., Ardlie K., Chan V., Myer V.E., Weber B.L., Porter J., Warmuth M., Finan P., Harris J.L., Meyerson M.L., Golub T.R., Morrissey M.P., Sellers W.R., Schlegel R., Garraway L.A.
The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity.
Nature 483:603-607(2012)
PubMed=23671654; DOI=10.1371/journal.pone.0063056
Lu Y.-H., Soong T.D., Elemento O.
A novel approach for characterizing microsatellite instability in cancer cells.
PLoS ONE 8:E63056-E63056(2013)
PubMed=25984343; DOI=10.1038/sdata.2014.35
Cowley G.S., Weir B.A., Vazquez F., Tamayo P., Scott J.A., Rusin S., East-Seletsky A., Ali L.D., Gerath W.F.J., Pantel S.E., Lizotte P.H., Jiang G.-Z., Hsiao J., Tsherniak A., Dwinell E., Aoyama S., Okamoto M., Harrington W., Gelfand E.T., Green T.M., Tomko M.J., Gopal S., Wong T.C., Li H.-B., Howell S., Stransky N., Liefeld T., Jang D., Bistline J., Meyers B.H., Armstrong S.A., Anderson K.C., Stegmaier K., Reich M., Pellman D., Boehm J.S., Mesirov J.P., Golub T.R., Root D.E., Hahn W.C.
Parallel genome-scale loss of function screens in 216 cancer cell lines for the identification of context-specific genetic dependencies.
Sci. Data 1:140035-140035(2014)
PubMed=25877200; DOI=10.1038/nature14397
Yu M., Selvaraj S.K., Liang-Chu M.M.Y., Aghajani S., Busse M., Yuan J., Lee G., Peale F.V., Klijn C., Bourgon R., Kaminker J.S., Neve R.M.
A resource for cell line authentication, annotation and quality control.
Nature 520:307-311(2015)
PubMed=26589293; DOI=10.1186/s13073-015-0240-5
Scholtalbers J., Boegel S., Bukur T., Byl M., Goerges S., Sorn P., Loewer M., Sahin U., Castle J.C.
TCLP: an online cancer cell line catalogue integrating HLA type, predicted neo-epitopes, virus and gene expression.
Genome Med. 7:118.1-118.7(2015)
PubMed=27397505; DOI=10.1016/j.cell.2016.06.017
Iorio F., Knijnenburg T.A., Vis D.J., Bignell G.R., Menden M.P., Schubert M., Aben N., Goncalves E., Barthorpe S., Lightfoot H., Cokelaer T., Greninger P., van Dyk E., Chang H., de Silva H., Heyn H., Deng X.-M., Egan R.K., Liu Q.-S., Mironenko T., Mitropoulos X., Richardson L., Wang J.-H., Zhang T.-H., Moran S., Sayols S., Soleimani M., Tamborero D., Lopez-Bigas N., Ross-Macdonald P., Esteller M., Gray N.S., Haber D.A., Stratton M.R., Benes C.H., Wessels L.F.A., Saez-Rodriguez J., McDermott U., Garnett M.J.
A landscape of pharmacogenomic interactions in cancer.
Cell 166:740-754(2016)
PubMed=30894373; DOI=10.1158/0008-5472.CAN-18-2747
Dutil J., Chen Z.-H., Monteiro A.N.A., Teer J.K., Eschrich S.A.
An interactive resource to probe genetic diversity and estimated ancestry in cancer cell lines.
Cancer Res. 79:1263-1273(2019)
PubMed=31068700; DOI=10.1038/s41586-019-1186-3
Ghandi M., Huang F.W., Jane-Valbuena J., Kryukov G.V., Lo C.C., McDonald E.R. III, Barretina J.G., Gelfand E.T., Bielski C.M., Li H.-X., Hu K., Andreev-Drakhlin A.Y., Kim J., Hess J.M., Haas B.J., Aguet F., Weir B.A., Rothberg M.V., Paolella B.R., Lawrence M.S., Akbani R., Lu Y.-L., Tiv H.L., Gokhale P.C., de Weck A., Mansour A.A., Oh C., Shih J., Hadi K., Rosen Y., Bistline J., Venkatesan K., Reddy A., Sonkin D., Liu M., Lehar J., Korn J.M., Porter D.A., Jones M.D., Golji J., Caponigro G., Taylor J.E., Dunning C.M., Creech A.L., Warren A.C., McFarland J.M., Zamanighomi M., Kauffmann A., Stransky N., Imielinski M., Maruvka Y.E., Cherniack A.D., Tsherniak A., Vazquez F., Jaffe J.D., Lane A.A., Weinstock D.M., Johannessen C.M., Morrissey M.P., Stegmeier F., Schlegel R., Hahn W.C., Getz G., Mills G.B., Boehm J.S., Golub T.R., Garraway L.A., Sellers W.R.
Next-generation characterization of the Cancer Cell Line Encyclopedia.
Nature 569:503-508(2019)
PubMed=35839778; DOI=10.1016/j.ccell.2022.06.010
Goncalves E., Poulos R.C., Cai Z.-X., Barthorpe S., Manda S.S., Lucas N., Beck A., Bucio-Noble D., Dausmann M., Hall C., Hecker M., Koh J., Lightfoot H., Mahboob S., Mali I., Morris J., Richardson L., Seneviratne A.J., Shepherd R., Sykes E., Thomas F., Valentini S., Williams S.G., Wu Y.-X., Xavier D., MacKenzie K.L., Hains P.G., Tully B., Robinson P.J., Zhong Q., Garnett M.J., Reddel R.R.
Pan-cancer proteomic map of 949 human cell lines.
Cancer Cell 40:835-849.e8(2022)