|
产品名称 |
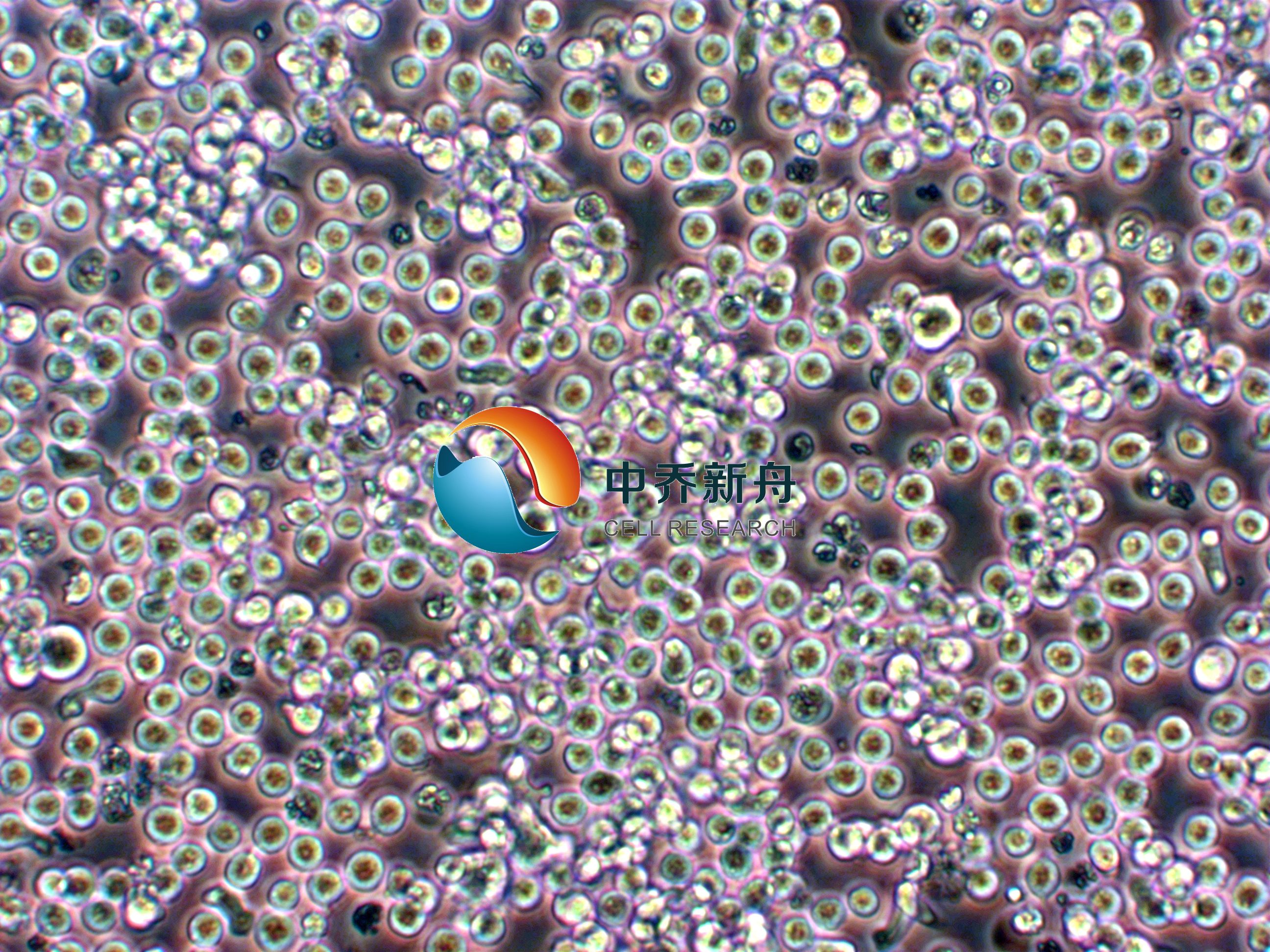
SUP-T1人T淋巴瘤细胞 |
|
货号 |
ZQ0839 |
|
产品介绍 |
该系来源于从患有 T 细胞淋巴细胞淋巴瘤的 8 岁儿童的恶性胸腔积液中收集的恶性细胞。细胞的形态为淋巴母细胞样,生长方式为悬浮生长。SUP-T1细胞系被广泛用于癌症研究,包括药物筛选、基因表达研究、细胞信号传导研究等。也用于研究T细胞急性淋巴细胞白血病(T-ALL)的发生、发展,以及Notch1基因表达对T-ALL细胞增殖及硼替佐米敏感性的影响。 |
|
种属 |
人 |
|
性别/年龄 |
男/8岁 |
|
组织 |
白血病;淋巴细胞;T 淋巴母细胞 |
|
疾病 |
淋巴母细胞淋巴瘤;T细胞 |
|
细胞类型 |
T淋巴母细胞 |
|
形态学 |
淋巴母细胞 |
|
生长方式 |
悬浮 |
|
倍增时间 |
大约28 hours (PubMed=3004618); ~30 hours (DSMZ=ACC-140); ~24-36 hours (HIVReagentProgram=ARP-100) |
|
培养基和添加剂 |
RPMI-1640(品牌:中乔新舟 货号:ZQ-200)+10%胎牛血清(中乔新舟 货号:ZQ0500)+1%P/S(中乔新舟 货号:CSP006) |
|
推荐完全培养基货号 |
|
|
生物安全等级 |
BSL-1 |
|
培养条件 |
95%空气,5%二氧化碳;37℃ |
|
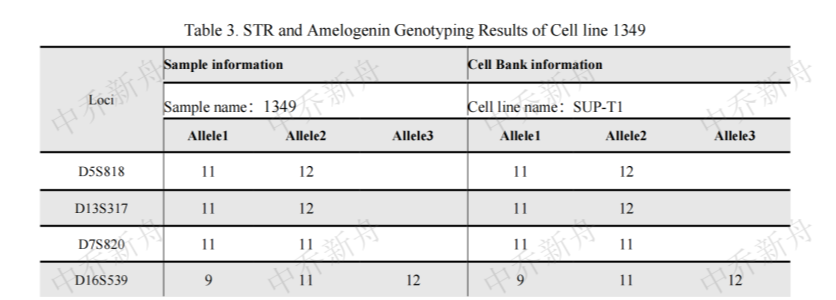
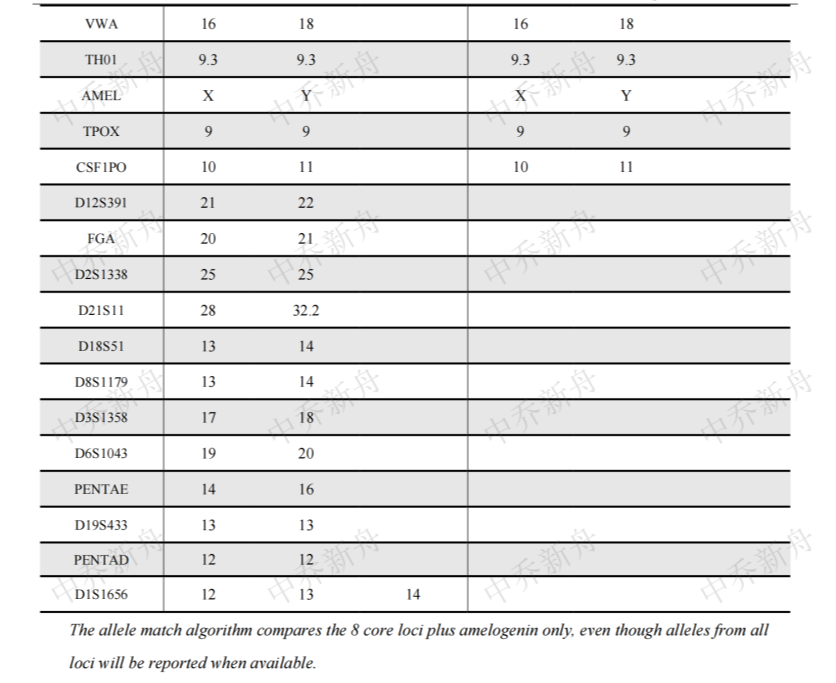
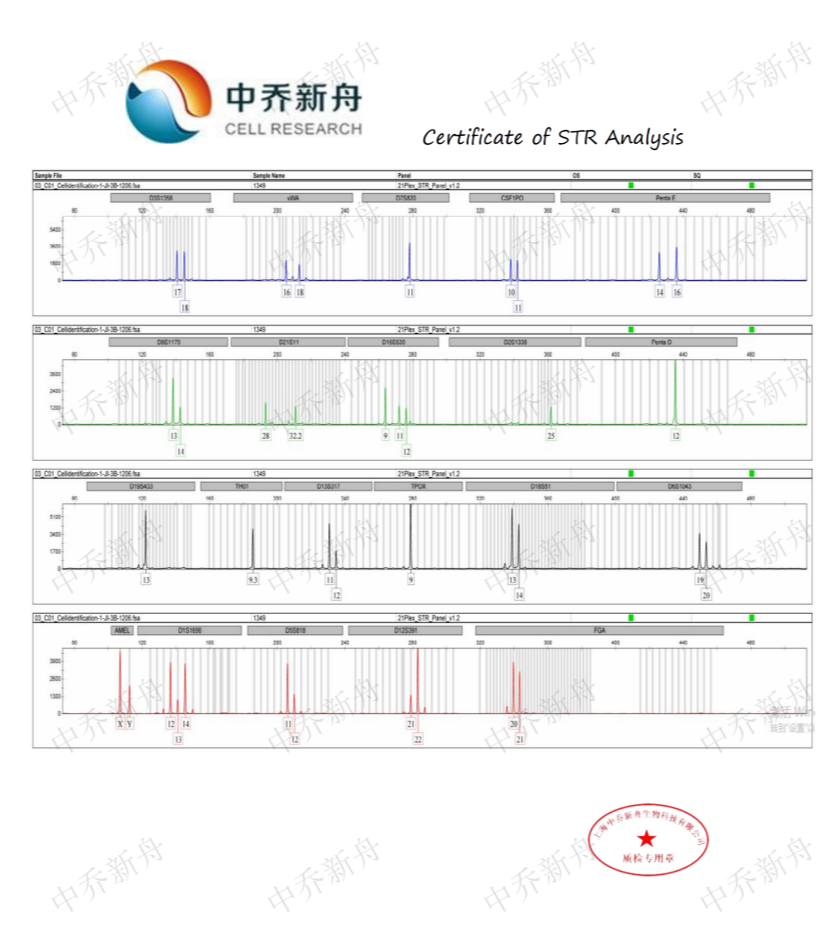
STR位点信息 |
Amelogenin :X (DSMZ=ACC-140) :X,Y (ATCC=CRL-1942; Cosmic-CLP=909743; PubMed=32938764) CSF1PO: 10,11 (ATCC=CRL-1942; Cosmic-CLP=909743; DSMZ=ACC-140) :10,11,12 (PubMed=32938764) D2S1338: 25,26 D3S1358: 16,17,18,19 (PubMed=32938764) :17 (DSMZ=ACC-140) D5S818 :10,11,12 (DSMZ=ACC-140) :11 (PubMed=32938764) :11,12 (ATCC=CRL-1942; Cosmic-CLP=909743) D7S820: 11 D8S1179: 13,14 (PubMed=32938764) :13,14,15 (DSMZ=ACC-140) D13S317: 10,11,12 (DSMZ=ACC-140; PubMed=32938764) :11,12 (ATCC=CRL-1942; Cosmic-CLP=909743) D16S539: 9,10,11,13 (DSMZ=ACC-140) :9,11,12 (ATCC=CRL-1942; Cosmic-CLP=909743) :9,12 (PubMed=32938764) D18S51 :13,14 D19S433: 13,27 D21S11 :28,29,32.2,33.2 (DSMZ=ACC-140) :28,31,32,33 (PubMed=32938764) FGA :19,20,21 (PubMed=32938764) :20,21 (DSMZ=ACC-140) Penta D: 12 Penta E :13,14,16 (PubMed=32938764) :14,16 (DSMZ=ACC-140) TH01: 9.3 TPOX :8,9 (DSMZ=ACC-140) :9 (ATCC=CRL-1942; Cosmic-CLP=909743; PubMed=32938764) vWA: 16,17,18,19 (PubMed=32938764) :16,18 (ATCC=CRL-1942; Cosmic-CLP=909743) |
|
抗原表达/受体表达 |
*** |
|
基因表达 |
*** |
|
保藏机构 |
ATCC |
|
供应限制 |
仅供科研使用 |
|
货号 |
ZQ0839 |
|
发货规格 |
活细胞:T25培养瓶*1瓶或者1ml 冻存管*2支(细胞量约为1x10^6 Cells/Vial)二选一 |
|
发货形式 |
活细胞:常温运输;冻存管:干冰运输 |
|
储存温度 |
活细胞:培养箱;冻存管:液氮罐 |
|
产地 |
中国 |
|
供应限制 |
仅供科研使用 |
PubMed=6437672
Smith S.D., Shatsky M., Cohen P.S., Warnke R.A., Link M.P., Glader B.E.
Monoclonal antibody and enzymatic profiles of human malignant T-lymphoid cells and derived cell lines.
Cancer Res. 44:5657-5660(1984)
PubMed=6438800; DOI=10.1126/science.6438800
Hecht F., Morgan R., Kaiser-McCaw Hecht B., Smith S.D.
Common region on chromosome 14 in T-cell leukemia and lymphoma.
Science 226:1445-1447(1984)
PubMed=3935328; DOI=10.1016/0092-8674(85)90243-0
Baer R., Chen K.-C., Smith S.D., Rabbitts T.H.
Fusion of an immunoglobulin variable gene and a T cell receptor constant gene in the chromosome 14 inversion associated with T cell tumors.
Cell 43:705-713(1985)
PubMed=3004618; DOI=10.1182/blood.V67.3.650.650
Smith S.D., Morgan R., Link M.P., McFall P., Hecht F.
Cytogenetic and immunophenotypic analysis of cell lines established from patients with T cell leukemia/lymphoma.
Blood 67:650-656(1986)
PubMed=3010463; DOI=10.1126/science.3010463
Lifson J.D., Reyes G.R., McGrath M.S., Stein B.S., Engleman E.G.
AIDS retrovirus induced cytopathology: giant cell formation and involvement of CD4 antigen.
Science 232:1123-1127(1986)
PubMed=3036367; DOI=10.1016/0092-8674(87)90666-0
Baer R., Forster A., Rabbitts T.H.
The mechanism of chromosome 14 inversion in a human T cell lymphoma.
Cell 50:97-105(1987)
PubMed=3107838; DOI=10.1016/0092-8674(87)90542-3
Stein B.S., Gowda S.D., Lifson J.D., Penhallow R.C., Bensch K.G., Engleman E.G.
pH-independent HIV entry into CD4-positive T cells via virus envelope fusion to the plasma membrane.
Cell 49:659-668(1987)
PubMed=7849311; DOI=10.1182/blood.V85.4.893.bloodjournal854893
Stranks G., Height S.E., Mitchell P., Jadayel D.M., Yuille M.A.R., De Lord C., Clutterbuck R.D., Treleaven J.G., Powles R.L., Nacheva E., Oscier D.G., Karpas A., Lenoir G.M., Smith S.D., Millar J.L., Catovsky D., Dyer M.J.S.
Deletions and rearrangement of CDKN2 in lymphoid malignancy.
Blood 85:893-901(1995)
PubMed=8558920
Dirks W.G., Zaborski M., Jager K., Challier C., Shiota M., Quentmeier H., Drexler H.G.
The (2;5)(p23;q35) translocation in cell lines derived from malignant lymphomas: absence of t(2;5) in Hodgkin-analogous cell lines.
Leukemia 10:142-149(1996)
PubMed=9933131; DOI=10.1016/S0145-2126(98)00133-7
Burger R., Hansen-Hagge T.E., Drexler H.G., Gramatzki M.
Heterogeneity of T-acute lymphoblastic leukemia (T-ALL) cell lines: suggestion for classification by immunophenotype and T-cell receptor studies.
Leuk. Res. 23:19-27(1999)
DOI=10.1016/B978-0-12-221970-2.50457-5
Drexler H.G.
The leukemia-lymphoma cell line factsbook.
(In) ISBN 9780122219702; pp.1-733; Academic Press; London (2001)
PubMed=11264379; DOI=10.1128/JVI.75.8.3903-3915.2001
Means R.E., Matthews T., Hoxie J.A., Malim M.H., Kodama T., Desrosiers R.C.
Ability of the V3 loop of simian immunodeficiency virus to serve as a target for antibody-mediated neutralization: correlation of neutralization sensitivity, growth in macrophages, and decreased dependence on CD4.
J. Virol. 75:3903-3915(2001)
PubMed=17170727; DOI=10.1038/sj.leu.2404486
Sandberg Y., Verhaaf B., van Gastel-Mol E.J., Wolvers-Tettero I.L.M., De Vos J., MacLeod R.A.F., Noordzij J.G., Dik W.A., van Dongen J.J.M., Langerak A.W.
Human T-cell lines with well-defined T-cell receptor gene rearrangements as controls for the BIOMED-2 multiplex polymerase chain reaction tubes.
Leukemia 21:230-237(2007)
PubMed=20164919; DOI=10.1038/nature08768
Bignell G.R., Greenman C.D., Davies H., Butler A.P., Edkins S., Andrews J.M., Buck G., Chen L., Beare D., Latimer C., Widaa S., Hinton J., Fahey C., Fu B.-Y., Swamy S., Dalgliesh G.L., Teh B.T., Deloukas P., Yang F.-T., Campbell P.J., Futreal P.A., Stratton M.R.
Signatures of mutation and selection in the cancer genome.
Nature 463:893-898(2010)
PubMed=22292511; DOI=10.2174/187221512799303172
Ho P., Dai K.-S., Chen H.-L.
Molecular cloning of a novel PPEF-1 gene variant from a T-cell lymphoblastic lymphoma cell line.
Recent Pat. DNA Gene Seq. 6:72-77(2012)
PubMed=22460905; DOI=10.1038/nature11003
Barretina J.G., Caponigro G., Stransky N., Venkatesan K., Margolin A.A., Kim S., Wilson C.J., Lehar J., Kryukov G.V., Sonkin D., Reddy A., Liu M., Murray L., Berger M.F., Monahan J.E., Morais P., Meltzer J., Korejwa A., Jane-Valbuena J., Mapa F.A., Thibault J., Bric-Furlong E., Raman P., Shipway A., Engels I.H., Cheng J., Yu G.-Y.K., Yu J.-J., Aspesi P. Jr., de Silva M., Jagtap K., Jones M.D., Wang L., Hatton C., Palescandolo E., Gupta S., Mahan S., Sougnez C., Onofrio R.C., Liefeld T., MacConaill L.E., Winckler W., Reich M., Li N.-X., Mesirov J.P., Gabriel S.B., Getz G., Ardlie K., Chan V., Myer V.E., Weber B.L., Porter J., Warmuth M., Finan P., Harris J.L., Meyerson M.L., Golub T.R., Morrissey M.P., Sellers W.R., Schlegel R., Garraway L.A.
The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity.
Nature 483:603-607(2012)
PubMed=22675565; DOI=10.1371/journal.pone.0038463
Atak Z.K., De Keersmaecker K., Gianfelici V., Geerdens E., Vandepoel R., Pauwels D., Porcu M., Lahortiga I., Brys V., Dirks W.G., Quentmeier H., Cloos J., Cuppens H., Uyttebroeck A., Vandenberghe P., Cools J., Aerts S.
High accuracy mutation detection in leukemia on a selected panel of cancer genes.
PLoS ONE 7:E38463-E38463(2012)
PubMed=25355872; DOI=10.1128/JVI.02570-14
Cao S.-B., Strong M.J., Wang X., Moss W.N., Concha M., Lin Z., O'Grady T., Baddoo M., Fewell C., Renne R., Flemington E.K.
High-throughput RNA sequencing-based virome analysis of 50 lymphoma cell lines from the Cancer Cell Line Encyclopedia project.
J. Virol. 89:713-729(2015)
PubMed=27397505; DOI=10.1016/j.cell.2016.06.017
Iorio F., Knijnenburg T.A., Vis D.J., Bignell G.R., Menden M.P., Schubert M., Aben N., Goncalves E., Barthorpe S., Lightfoot H., Cokelaer T., Greninger P., van Dyk E., Chang H., de Silva H., Heyn H., Deng X.-M., Egan R.K., Liu Q.-S., Mironenko T., Mitropoulos X., Richardson L., Wang J.-H., Zhang T.-H., Moran S., Sayols S., Soleimani M., Tamborero D., Lopez-Bigas N., Ross-Macdonald P., Esteller M., Gray N.S., Haber D.A., Stratton M.R., Benes C.H., Wessels L.F.A., Saez-Rodriguez J., McDermott U., Garnett M.J.
A landscape of pharmacogenomic interactions in cancer.
Cell 166:740-754(2016)
PubMed=29304865; DOI=10.1186/s12985-017-0917-z
Becerra-Artiles A., Santoro T., Stern L.J.
Evaluation of a method to measure HHV-6B infection in vitro based on cell size.
Virol. J. 15:4.1-4.15(2018)
PubMed=30285677; DOI=10.1186/s12885-018-4840-5
Tan K.-T., Ding L.-W., Sun Q.-Y., Lao Z.-T., Chien W., Ren X., Xiao J.-F., Loh X.-Y., Xu L., Lill M., Mayakonda A., Lin D.-C., Yang H., Koeffler H.P.
Profiling the B/T cell receptor repertoire of lymphocyte derived cell lines.
BMC Cancer 18:940.1-940.13(2018)
PubMed=30629668; DOI=10.1371/journal.pone.0210404
Uphoff C.C., Pommerenke C., Denkmann S.A., Drexler H.G.
Screening human cell lines for viral infections applying RNA-Seq data analysis.
PLoS ONE 14:E0210404-E0210404(2019)
PubMed=30894373; DOI=10.1158/0008-5472.CAN-18-2747
Dutil J., Chen Z.-H., Monteiro A.N.A., Teer J.K., Eschrich S.A.
An interactive resource to probe genetic diversity and estimated ancestry in cancer cell lines.
Cancer Res. 79:1263-1273(2019)
PubMed=31068700; DOI=10.1038/s41586-019-1186-3
Ghandi M., Huang F.W., Jane-Valbuena J., Kryukov G.V., Lo C.C., McDonald E.R. III, Barretina J.G., Gelfand E.T., Bielski C.M., Li H.-X., Hu K., Andreev-Drakhlin A.Y., Kim J., Hess J.M., Haas B.J., Aguet F., Weir B.A., Rothberg M.V., Paolella B.R., Lawrence M.S., Akbani R., Lu Y.-L., Tiv H.L., Gokhale P.C., de Weck A., Mansour A.A., Oh C., Shih J., Hadi K., Rosen Y., Bistline J., Venkatesan K., Reddy A., Sonkin D., Liu M., Lehar J., Korn J.M., Porter D.A., Jones M.D., Golji J., Caponigro G., Taylor J.E., Dunning C.M., Creech A.L., Warren A.C., McFarland J.M., Zamanighomi M., Kauffmann A., Stransky N., Imielinski M., Maruvka Y.E., Cherniack A.D., Tsherniak A., Vazquez F., Jaffe J.D., Lane A.A., Weinstock D.M., Johannessen C.M., Morrissey M.P., Stegmeier F., Schlegel R., Hahn W.C., Getz G., Mills G.B., Boehm J.S., Golub T.R., Garraway L.A., Sellers W.R.
Next-generation characterization of the Cancer Cell Line Encyclopedia.
Nature 569:503-508(2019)
PubMed=32938764; DOI=10.1128/JVI.01334-20
Fernandez M.V., Hoffman H.K., Pezeshkian N., Tedbury P.R., van Engelenburg S.B., Freed E.O.
Elucidating the basis for permissivity of the MT-4 T-cell line to replication of an HIV-1 mutant lacking the gp41 cytoplasmic tail.
J. Virol. 94:e01334.20.1-e01334.20.27(2020)
PubMed=35839778; DOI=10.1016/j.ccell.2022.06.010
Goncalves E., Poulos R.C., Cai Z.-X., Barthorpe S., Manda S.S., Lucas N., Beck A., Bucio-Noble D., Dausmann M., Hall C., Hecker M., Koh J., Lightfoot H., Mahboob S., Mali I., Morris J., Richardson L., Seneviratne A.J., Shepherd R., Sykes E., Thomas F., Valentini S., Williams S.G., Wu Y.-X., Xavier D., MacKenzie K.L., Hains P.G., Tully B., Robinson P.J., Zhong Q., Garnett M.J., Reddel R.R.
Pan-cancer proteomic map of 949 human cell lines.
Cancer Cell 40:835-849.e8(2022)
 上海中乔新舟生物科技有限公司
上海中乔新舟生物科技有限公司