|
产品名称 |
TOV21G人卵巢癌细胞 |
|
货号 |
ZQ1008 |
|
产品介绍 |
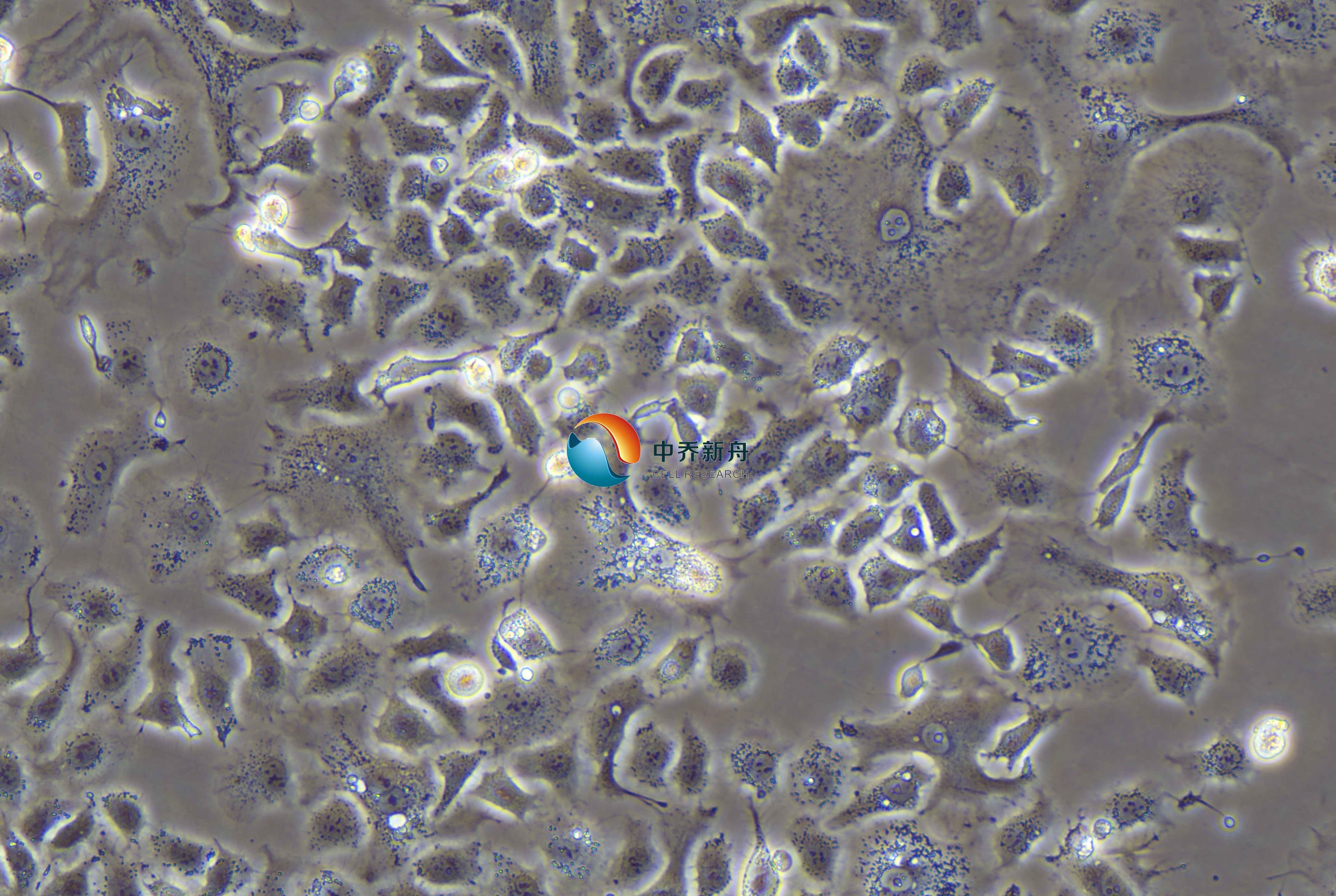
TOV21G是一种人卵巢癌细胞系,它源自一位62岁法裔加拿大女性的原发性恶性腺癌,于 1991 年 10 月建系。该患者没有卵巢癌的家族史。这种细胞系在生物医学研究中被广泛使用,尤其是在研究卵巢癌的生物学特性、病理机制以及药物筛选等方面。该系在染色体 3p24 处未显示缺失。 |
|
种属 |
人 |
|
性别/年龄 |
女/62岁 |
|
组织 |
卵巢 |
|
疾病 |
腺癌 |
|
细胞类型 |
肿瘤细胞 |
|
形态学 |
上皮样 |
|
生长方式 |
贴壁 |
|
倍增时间 |
1.5 days (PubMed=10949993); 27 hours (PubMed=25984343); 30.62 hours (JWGray panel) |
|
培养基和添加剂 |
MCDB105+Medium199(1:1)+15%FBS(货号:ZQ0500)+1%PS(中乔新舟 货号:CSP006) |
|
推荐完全培养基货号 |
|
|
生物安全等级 |
BSL-1 |
|
培养条件 |
95%空气,5%二氧化碳;37℃ |
|
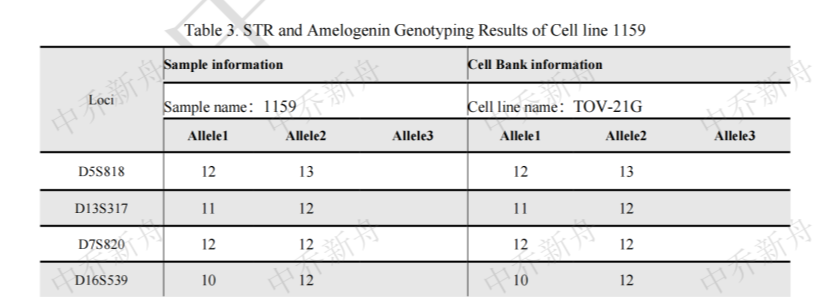
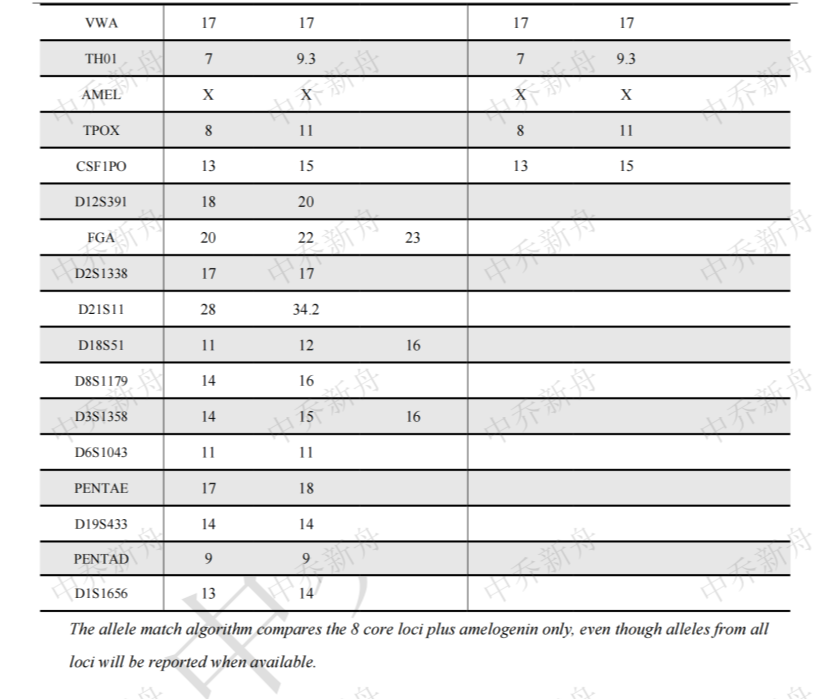
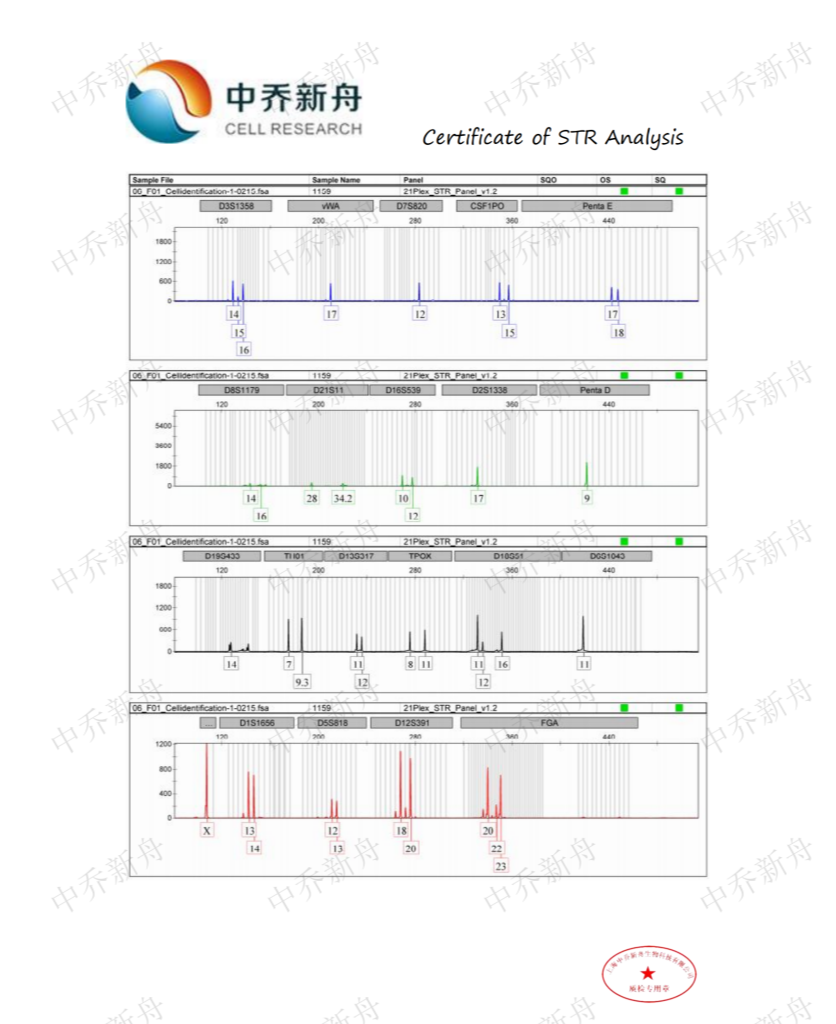
STR位点信息 |
Amelogenin :X CSF1PO: 13,15 D2S1338: 17 D3S1358: 14,16 D5S818: 12,13 D7S820 :12 D8S1179 :13,16 D13S317 :11,12 D16S539: 10,12 D18S51: 12 (PubMed=25230021; PubMed=25877200) :12,16 (PubMed=22710073) D19S433: 14,16.2 D21S11: 28,34.2 FGA: 20,23 Penta D :9 Penta E: 17,18 (PubMed=25877200) :17,19 (PubMed=25230021) TH01: 7,9.3 TPOX :8,11 vWA :17 (ATCC=CRL-3577; Cosmic-CLP=1240222; PubMed=22710073; PubMed=25230021; PubMed=25877200; PubMed=30485824) :17,18 (Direct_author_submission) |
|
抗原表达/受体表达 |
*** |
|
基因表达 |
*** |
|
保藏机构 |
ATCC; CRL-11730;BCRC; 60407 |
|
供应限制 |
仅供科研使用 |
|
货号 |
ZQ1008 |
|
发货规格 |
活细胞:T25培养瓶*1瓶或者1ml 冻存管*2支(细胞量约为1x10^6 Cells/Vial )二选一 |
|
发货形式 |
活细胞:常温运输;冻存管:干冰运输 |
|
储存温度 |
活细胞:培养箱;冻存管:液氮罐 |
|
产地 |
中国 |
|
供应限制 |
仅供科研使用 |
Patent=US5710038
Mes-Masson A.-M., Provencher D.M.
Primary cultures of normal and tumoral human ovarian epithelium.
Patent number US5710038, 20-Jan-1998
PubMed=10949993; DOI=10.1290/1071-2690(2000)036<0357:COFNEO>2.0.CO;2
Provencher D.M., Lounis H., Champoux L., Tetrault M., Manderson E.N., Wang J.-C., Eydoux P., Savoie R., Tonin P.N., Mes-Masson A.-M.
Characterization of four novel epithelial ovarian cancer cell lines.
In Vitro Cell. Dev. Biol. Anim. 36:357-361(2000)
PubMed=11641787; DOI=10.1038/sj.onc.1204804
Tonin P.N., Hudson T.J., Rodier F., Bossolasco M., Lee P.D., Novak J., Manderson E.N., Provencher D.M., Mes-Masson A.-M.
Microarray analysis of gene expression mirrors the biology of an ovarian cancer model.
Oncogene 20:6617-6626(2001)
PubMed=19058220; DOI=10.1002/ijc.24058
Dafou D., Ramus S.J., Choi K., Grun B., Trott D.A., Newbold R.F., Jacobs I.J., Jones C., Gayther S.A.
Chromosomes 6 and 18 induce neoplastic suppression in epithelial ovarian cancer cells.
Int. J. Cancer 124:1037-1044(2009)
PubMed=20081105; DOI=10.1210/me.2009-0295
Nagaraja A.K., Creighton C.J., Yu Z.-F., Zhu H.-F., Gunaratne P.H., Reid J.G., Olokpa E., Itamochi H., Ueno N.T., Hawkins S.M., Anderson M.L., Matzuk M.M.
A link between mir-100 and FRAP1/mTOR in clear cell ovarian cancer.
Mol. Endocrinol. 24:447-463(2010)
PubMed=20204287; DOI=10.3892/or_00000728
Langland G.T., Yannone S.M., Langland R.A., Nakao A., Guan Y.-H., Long S.B.T., Vonguyen L., Chen D.J., Gray J.W., Chen F.-Q.
Radiosensitivity profiles from a panel of ovarian cancer cell lines exhibiting genetic alterations in p53 and disparate DNA-dependent protein kinase activities.
Oncol. Rep. 23:1021-1026(2010)
PubMed=22460905; DOI=10.1038/nature11003
Barretina J.G., Caponigro G., Stransky N., Venkatesan K., Margolin A.A., Kim S., Wilson C.J., Lehar J., Kryukov G.V., Sonkin D., Reddy A., Liu M., Murray L., Berger M.F., Monahan J.E., Morais P., Meltzer J., Korejwa A., Jane-Valbuena J., Mapa F.A., Thibault J., Bric-Furlong E., Raman P., Shipway A., Engels I.H., Cheng J., Yu G.-Y.K., Yu J.-J., Aspesi P. Jr., de Silva M., Jagtap K., Jones M.D., Wang L., Hatton C., Palescandolo E., Gupta S., Mahan S., Sougnez C., Onofrio R.C., Liefeld T., MacConaill L.E., Winckler W., Reich M., Li N.-X., Mesirov J.P., Gabriel S.B., Getz G., Ardlie K., Chan V., Myer V.E., Weber B.L., Porter J., Warmuth M., Finan P., Harris J.L., Meyerson M.L., Golub T.R., Morrissey M.P., Sellers W.R., Schlegel R., Garraway L.A.
The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity.
Nature 483:603-607(2012)
PubMed=22705003; DOI=10.1016/j.humpath.2012.03.011
Rahman M., Nakayama K., Rahman M.T., Nakayama N., Ishikawa M., Katagiri A., Iida K., Nakayama S., Otsuki Y., Shih I.-M., Miyazaki K.
Clinicopathologic and biological analysis of PIK3CA mutation in ovarian clear cell carcinoma.
Hum. Pathol. 43:2197-2206(2012)
PubMed=22710073; DOI=10.1016/j.ygyno.2012.06.017
Korch C.T., Spillman M.A., Jackson T.A., Jacobsen B.M., Murphy S.K., Lessey B.A., Jordan V.C., Bradford A.P.
DNA profiling analysis of endometrial and ovarian cell lines reveals misidentification, redundancy and contamination.
Gynecol. Oncol. 127:241-248(2012)
PubMed=23839242; DOI=10.1038/ncomms3126
Domcke S., Sinha R., Levine D.A., Sander C., Schultz N.
Evaluating cell lines as tumour models by comparison of genomic profiles.
Nat. Commun. 4:2126.1-2126.10(2013)
PubMed=24023729; DOI=10.1371/journal.pone.0072162
Anglesio M.S., Wiegand K.C., Melnyk N., Chow C., Salamanca C.M., Prentice L.M., Senz J., Yang W., Spillman M.A., Cochrane D.R., Shumansky K., Shah S.P., Kalloger S.E., Huntsman D.G.
Type-specific cell line models for type-specific ovarian cancer research.
PLoS ONE 8:E72162-E72162(2013)
PubMed=24224046; DOI=10.1371/journal.pone.0080229
Zanaruddin S.N.S., Yee P.-S., Hor S.Y., Kong Y.H., Ghani W.M.N.W.A., Mustafa W.M.W., Zain R.B., Prime S.S., Rahman Z.A.A., Cheong S.-C.
Common oncogenic mutations are infrequent in oral squamous cell carcinoma of Asian origin.
PLoS ONE 8:E80229-E80229(2013)
PubMed=25230021; DOI=10.1371/journal.pone.0103988
Beaufort C.M., Helmijr J.C.A., Piskorz A.M., Hoogstraat M., Ruigrok-Ritstier K., Besselink N., Murtaza M., van IJcken W.F.J., Heine A.A.J., Smid M., Koudijs M.J., Brenton J.D., Berns E.M.J.J., Helleman J.
Ovarian cancer cell line panel (OCCP): clinical importance of in vitro morphological subtypes.
PLoS ONE 9:E103988-E103988(2014)
PubMed=25960936; DOI=10.4161/21624011.2014.954893
Boegel S., Lower M., Bukur T., Sahin U., Castle J.C.
A catalog of HLA type, HLA expression, and neo-epitope candidates in human cancer cell lines.
OncoImmunology 3:e954893.1-e954893.12(2014)
PubMed=25984343; DOI=10.1038/sdata.2014.35
Cowley G.S., Weir B.A., Vazquez F., Tamayo P., Scott J.A., Rusin S., East-Seletsky A., Ali L.D., Gerath W.F.J., Pantel S.E., Lizotte P.H., Jiang G.-Z., Hsiao J., Tsherniak A., Dwinell E., Aoyama S., Okamoto M., Harrington W., Gelfand E.T., Green T.M., Tomko M.J., Gopal S., Wong T.C., Li H.-B., Howell S., Stransky N., Liefeld T., Jang D., Bistline J., Meyers B.H., Armstrong S.A., Anderson K.C., Stegmaier K., Reich M., Pellman D., Boehm J.S., Mesirov J.P., Golub T.R., Root D.E., Hahn W.C.
Parallel genome-scale loss of function screens in 216 cancer cell lines for the identification of context-specific genetic dependencies.
Sci. Data 1:140035-140035(2014)
PubMed=25877200; DOI=10.1038/nature14397
Yu M., Selvaraj S.K., Liang-Chu M.M.Y., Aghajani S., Busse M., Yuan J., Lee G., Peale F.V., Klijn C., Bourgon R., Kaminker J.S., Neve R.M.
A resource for cell line authentication, annotation and quality control.
Nature 520:307-311(2015)
PubMed=26388441; DOI=10.1016/j.cell.2015.08.056
Creixell P., Schoof E.M., Simpson C.D., Longden J., Miller C.J., Lou H.J., Perryman L., Cox T.R., Zivanovic N., Palmeri A., Wesolowska-Andersen A., Helmer-Citterich M., Ferkinghoff-Borg J., Itamochi H., Bodenmiller B., Erler J.T., Turk B.E., Linding R.
Kinome-wide decoding of network-attacking mutations rewiring cancer signaling.
Cell 163:202-217(2015)
PubMed=26589293; DOI=10.1186/s13073-015-0240-5
Scholtalbers J., Boegel S., Bukur T., Byl M., Goerges S., Sorn P., Loewer M., Sahin U., Castle J.C.
TCLP: an online cancer cell line catalogue integrating HLA type, predicted neo-epitopes, virus and gene expression.
Genome Med. 7:118.1-118.7(2015)
PubMed=27397505; DOI=10.1016/j.cell.2016.06.017
Iorio F., Knijnenburg T.A., Vis D.J., Bignell G.R., Menden M.P., Schubert M., Aben N., Goncalves E., Barthorpe S., Lightfoot H., Cokelaer T., Greninger P., van Dyk E., Chang H., de Silva H., Heyn H., Deng X.-M., Egan R.K., Liu Q.-S., Mironenko T., Mitropoulos X., Richardson L., Wang J.-H., Zhang T.-H., Moran S., Sayols S., Soleimani M., Tamborero D., Lopez-Bigas N., Ross-Macdonald P., Esteller M., Gray N.S., Haber D.A., Stratton M.R., Benes C.H., Wessels L.F.A., Saez-Rodriguez J., McDermott U., Garnett M.J.
A landscape of pharmacogenomic interactions in cancer.
Cell 166:740-754(2016)
PubMed=28196595; DOI=10.1016/j.ccell.2017.01.005
Li J., Zhao W., Akbani R., Liu W.-B., Ju Z.-L., Ling S.-Y., Vellano C.P., Roebuck P., Yu Q.-H., Eterovic A.K., Byers L.A., Davies M.A., Deng W.-L., Gopal Y.N.V., Chen G., von Euw E.M., Slamon D.J., Conklin D., Heymach J.V., Gazdar A.F., Minna J.D., Myers J.N., Lu Y.-L., Mills G.B., Liang H.
Characterization of human cancer cell lines by reverse-phase protein arrays.
Cancer Cell 31:225-239(2017)
PubMed=28273451; DOI=10.1016/j.celrep.2017.02.028
Medrano M., Communal L., Brown K.R., Iwanicki M., Normand J., Paterson J., Sircoulomb F., Krzyzanowski P.M., Novak M., Doodnauth S.A., Suarez Saiz F.J., Cullis J., Al-awar R., Neel B.G., McPherson J., Drapkin R.I., Ailles L., Mes-Masson A.-M., Rottapel R.
Interrogation of functional cell-surface markers identifies CD151 dependency in high-grade serous ovarian cancer.
Cell Rep. 18:2343-2358(2017)
PubMed=30485824; DOI=10.1016/j.celrep.2018.10.096
Papp E., Hallberg D., Konecny G.E., Bruhm D.C., Adleff V., Noe M., Kagiampakis I., Palsgrove D., Conklin D., Kinose Y., White J.R., Press M.F., Drapkin R.I., Easwaran H., Baylin S.B., Slamon D.J., Velculescu V.E., Scharpf R.B.
Integrated genomic, epigenomic, and expression analyses of ovarian cancer cell lines.
Cell Rep. 25:2617-2633(2018)
PubMed=30894373; DOI=10.1158/0008-5472.CAN-18-2747
Dutil J., Chen Z.-H., Monteiro A.N.A., Teer J.K., Eschrich S.A.
An interactive resource to probe genetic diversity and estimated ancestry in cancer cell lines.
Cancer Res. 79:1263-1273(2019)
PubMed=30971826; DOI=10.1038/s41586-019-1103-9
Behan F.M., Iorio F., Picco G., Goncalves E., Beaver C.M., Migliardi G., Santos R., Rao Y., Sassi F., Pinnelli M., Ansari R., Harper S., Jackson D.A., McRae R., Pooley R., Wilkinson P., van der Meer D.J., Dow D., Buser-Doepner C.A., Bertotti A., Trusolino L., Stronach E.A., Saez-Rodriguez J., Yusa K., Garnett M.J.
Prioritization of cancer therapeutic targets using CRISPR-Cas9 screens.
Nature 568:511-516(2019)
PubMed=31068700; DOI=10.1038/s41586-019-1186-3
Ghandi M., Huang F.W., Jane-Valbuena J., Kryukov G.V., Lo C.C., McDonald E.R. III, Barretina J.G., Gelfand E.T., Bielski C.M., Li H.-X., Hu K., Andreev-Drakhlin A.Y., Kim J., Hess J.M., Haas B.J., Aguet F., Weir B.A., Rothberg M.V., Paolella B.R., Lawrence M.S., Akbani R., Lu Y.-L., Tiv H.L., Gokhale P.C., de Weck A., Mansour A.A., Oh C., Shih J., Hadi K., Rosen Y., Bistline J., Venkatesan K., Reddy A., Sonkin D., Liu M., Lehar J., Korn J.M., Porter D.A., Jones M.D., Golji J., Caponigro G., Taylor J.E., Dunning C.M., Creech A.L., Warren A.C., McFarland J.M., Zamanighomi M., Kauffmann A., Stransky N., Imielinski M., Maruvka Y.E., Cherniack A.D., Tsherniak A., Vazquez F., Jaffe J.D., Lane A.A., Weinstock D.M., Johannessen C.M., Morrissey M.P., Stegmeier F., Schlegel R., Hahn W.C., Getz G., Mills G.B., Boehm J.S., Golub T.R., Garraway L.A., Sellers W.R.
Next-generation characterization of the Cancer Cell Line Encyclopedia.
Nature 569:503-508(2019)
PubMed=35839778; DOI=10.1016/j.ccell.2022.06.010
Goncalves E., Poulos R.C., Cai Z.-X., Barthorpe S., Manda S.S., Lucas N., Beck A., Bucio-Noble D., Dausmann M., Hall C., Hecker M., Koh J., Lightfoot H., Mahboob S., Mali I., Morris J., Richardson L., Seneviratne A.J., Shepherd R., Sykes E., Thomas F., Valentini S., Williams S.G., Wu Y.-X., Xavier D., MacKenzie K.L., Hains P.G., Tully B., Robinson P.J., Zhong Q., Garnett M.J., Reddel R.R.
Pan-cancer proteomic map of 949 human cell lines.
Cancer Cell 40:835-849.e8(2022)
 上海中乔新舟生物科技有限公司
上海中乔新舟生物科技有限公司