|
产品名称 |
LK-2人肺腺癌细胞 |
|
货号 |
ZQ1011 |
|
产品介绍 |
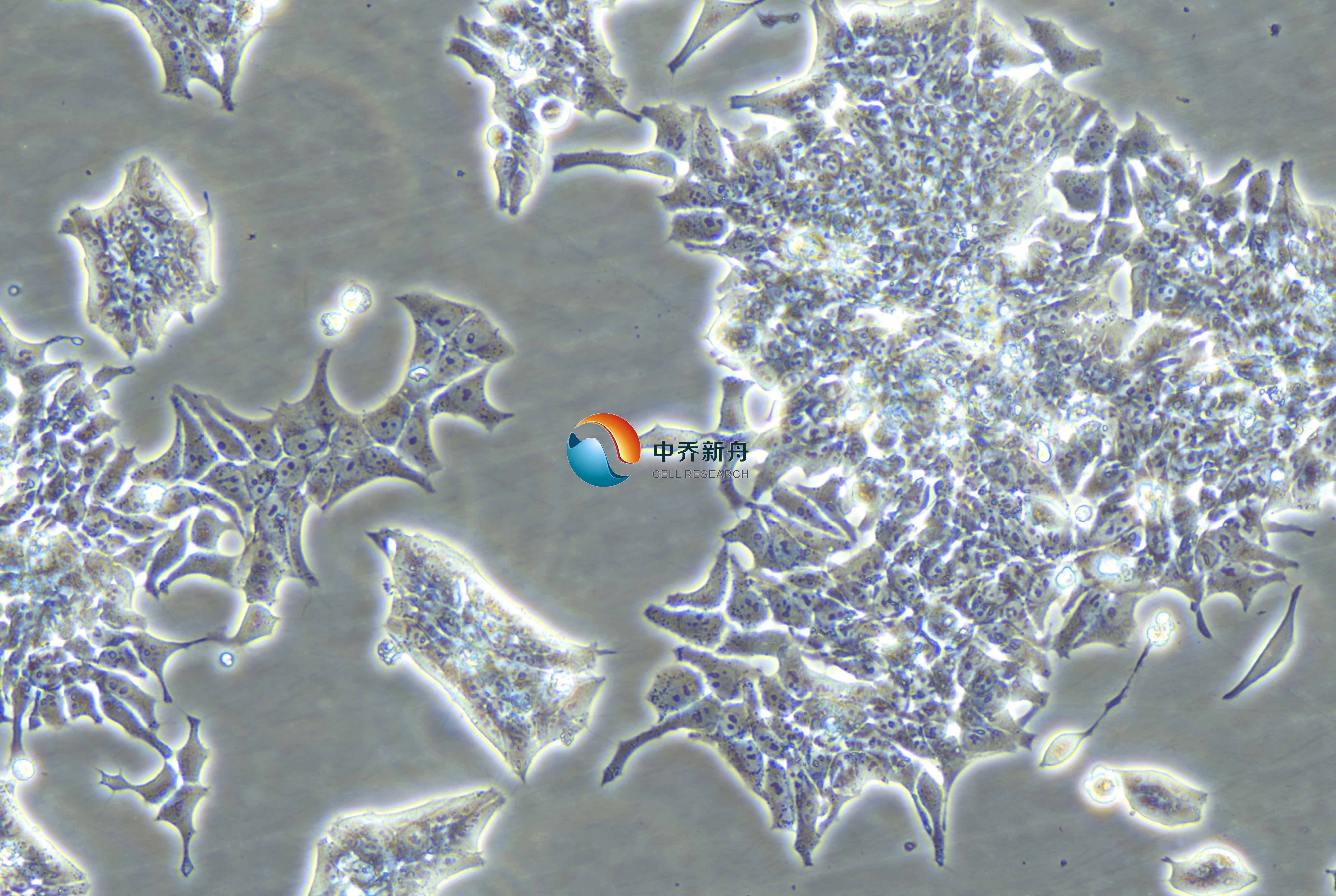
LK-2是一种人肺腺癌细胞系,它源自一位74岁男性的肺鳞状细胞癌组织。这种细胞系在生物医学研究中被广泛使用,尤其是在研究肺癌的生物学特性、病理机制以及药物筛选等方面。 |
|
种属 |
人 |
|
性别/年龄 |
男/74岁 |
|
组织 |
肺 |
|
疾病 |
鳞状细胞癌 |
|
细胞类型 |
肿瘤细胞 |
|
形态学 |
上皮样 |
|
生长方式 |
贴壁 |
|
倍增时间 |
32 hours (PubMed=25984343); ~27 hours (Note=Lot 04162009), ~26 hours (Note=Lot 11142016) (JCRB=JCRB0829) |
|
培养基和添加剂 |
RPMI-1640(品牌:中乔新舟 货号:ZQ-200)+10%胎牛血清(中乔新舟 货号:ZQ500-A)+1%P/S(中乔新舟 货号:CSP006) |
|
推荐完全培养基货号 |
|
|
生物安全等级 |
BSL-1 |
|
培养条件 |
95%空气,5%二氧化碳;37℃ |
|
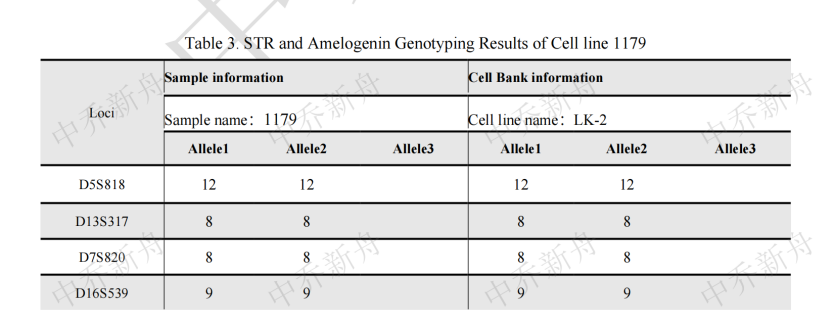
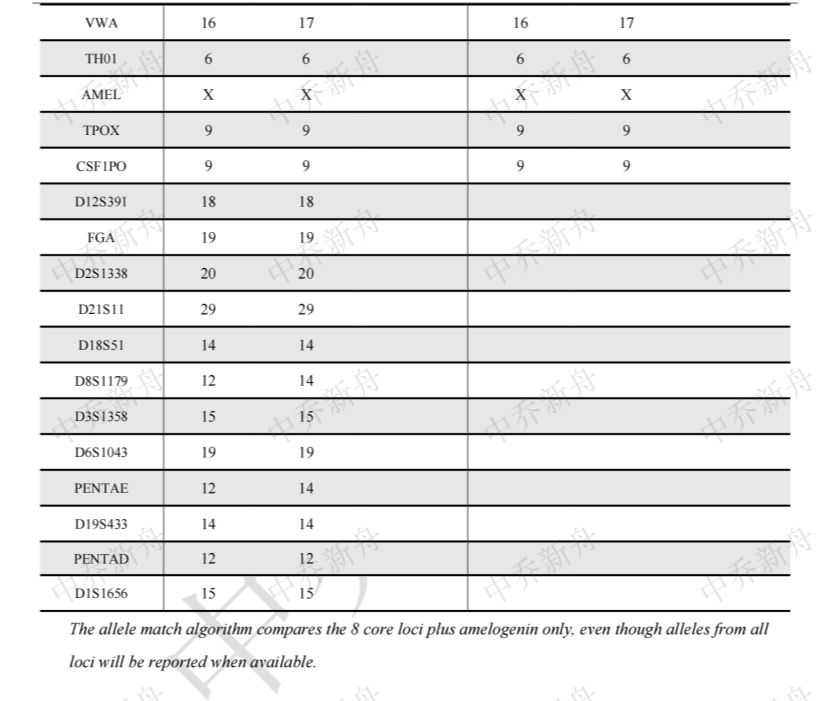
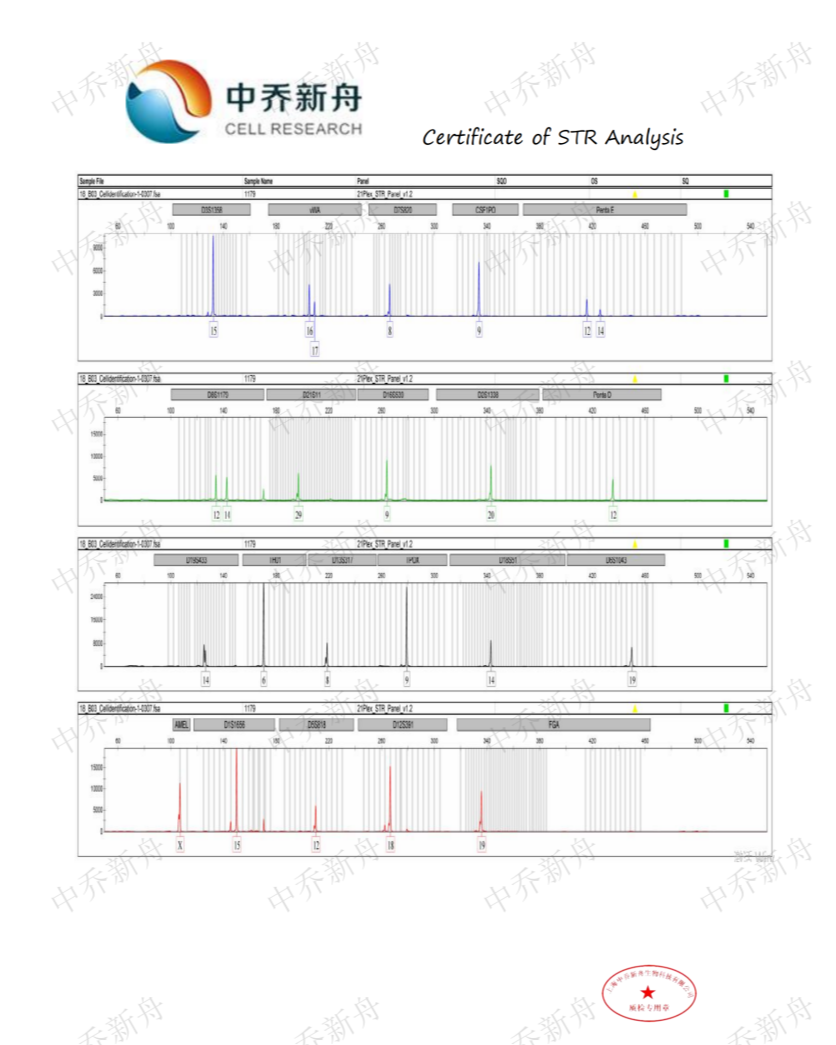
STR位点信息 |
Amelogenin: X CSF1PO: 9 D5S818 :12 (Cosmic-CLP=687787; JCRB=JCRB0829; RCB=RCB1970) :12,15 (TKG=TKG 0494) D7S820: 8 D13S317: 8 D16S539: 9 TH01: 6 TPOX: 9 vWA :16,17 |
|
抗原表达/受体表达 |
*** |
|
基因表达 |
*** |
|
保藏机构 |
JCRB; JCRB0829 |
|
供应限制 |
仅供科研使用 |
|
货号 |
ZQ1011 |
|
发货规格 |
活细胞:T25培养瓶*1瓶或者1ml 冻存管*2支(细胞量约为1x10^6 Cells/Vial )二选一 |
|
发货形式 |
活细胞:常温运输;冻存管:干冰运输 |
|
储存温度 |
活细胞:培养箱;冻存管:液氮罐 |
|
产地 |
中国 |
|
供应限制 |
仅供科研使用 |
CLPUB00586
Yoshioka S.
Studies on thiol protease inhibitor isolated from human lung cancer cell line.
Hiroshima J. Med. Sci. 37:199-215(1989)
PubMed=9290701; DOI=10.1002/(SICI)1098-2744(199708)19:4<243::AID-MC5>3.0.CO;2-D
Jia L.-Q., Osada M., Ishioka C., Gamo M., Ikawa S., Suzuki T., Shimodaira H., Niitani T., Kudo T., Akiyama M., Kimura N., Matsuo M., Mizusawa H., Tanaka N., Koyama H., Namba M., Kanamaru R., Kuroki T.
Screening the p53 status of human cell lines using a yeast functional assay.
Mol. Carcinog. 19:243-253(1997)
PubMed=10358721; DOI=10.18926/AMO/31626
Matsushita A., Tabata M., Ueoka H., Kiura K., Shibayama T., Aoe K., Kohara H., Harada M.
Establishment of a drug sensitivity panel using human lung cancer cell lines.
Acta Med. Okayama 53:67-75(1999)
PubMed=14581340
Yokoi S., Yasui K., Iizasa T., Imoto I., Fujisawa T., Inazawa J.
TERC identified as a probable target within the 3q26 amplicon that is detected frequently in non-small cell lung cancers.
Clin. Cancer Res. 9:4705-4713(2003)
PubMed=15746151; DOI=10.1093/hmg/ddi092
Izumi H., Inoue J., Yokoi S., Hosoda H., Shibata T., Sunamori M., Hirohashi S., Inazawa J., Imoto I.
Frequent silencing of DBC1 is by genetic or epigenetic mechanisms in non-small cell lung cancers.
Hum. Mol. Genet. 14:997-1007(2005)
PubMed=16105816; DOI=10.1158/0008-5472.CAN-05-0331
Nagai Y., Miyazawa H., Huqun X., Tanaka T., Udagawa K., Kato M., Fukuyama S., Yokote A., Kobayashi K., Kanazawa M., Hagiwara K.
Genetic heterogeneity of the epidermal growth factor receptor in non-small cell lung cancer cell lines revealed by a rapid and sensitive detection system, the peptide nucleic acid-locked nucleic acid PCR clamp.
Cancer Res. 65:7276-7282(2005)
PubMed=19472407; DOI=10.1002/humu.21028
Blanco R., Iwakawa R., Tang M.-Y., Kohno T., Angulo B., Pio R., Montuenga L.M., Minna J.D., Yokota J., Sanchez-Cespedes M.
A gene-alteration profile of human lung cancer cell lines.
Hum. Mutat. 30:1199-1206(2009)
PubMed=20164919; DOI=10.1038/nature08768
Bignell G.R., Greenman C.D., Davies H., Butler A.P., Edkins S., Andrews J.M., Buck G., Chen L., Beare D., Latimer C., Widaa S., Hinton J., Fahey C., Fu B.-Y., Swamy S., Dalgliesh G.L., Teh B.T., Deloukas P., Yang F.-T., Campbell P.J., Futreal P.A., Stratton M.R.
Signatures of mutation and selection in the cancer genome.
Nature 463:893-898(2010)
PubMed=20215515; DOI=10.1158/0008-5472.CAN-09-3458
Rothenberg S.M., Mohapatra G., Rivera M.N., Winokur D., Greninger P., Nitta M., Sadow P.M., Sooriyakumar G., Brannigan B.W., Ulman M.J., Perera R.M., Wang R., Tam A., Ma X.-J., Erlander M., Sgroi D.C., Rocco J.W., Lingen M.W., Cohen E.E.W., Louis D.N., Settleman J., Haber D.A.
A genome-wide screen for microdeletions reveals disruption of polarity complex genes in diverse human cancers.
Cancer Res. 70:2158-2164(2010)
PubMed=20557307; DOI=10.1111/j.1349-7006.2010.01622.x
Iwakawa R., Kohno T., Enari M., Kiyono T., Yokota J.
Prevalence of human papillomavirus 16/18/33 infection and p53 mutation in lung adenocarcinoma.
Cancer Sci. 101:1891-1896(2010)
PubMed=22313637; DOI=10.4161/cbt.19238
Takata M., Chikumi H., Miyake N., Adachi K., Kanamori Y., Yamasaki A., Igishi T., Burioka N., Nanba E., Shimizu E.
Lack of AKT activation in lung cancer cells with EGFR mutation is a novel marker of cetuximab sensitivity.
Cancer Biol. Ther. 13:369-378(2012)
PubMed=22460905; DOI=10.1038/nature11003
Barretina J.G., Caponigro G., Stransky N., Venkatesan K., Margolin A.A., Kim S., Wilson C.J., Lehar J., Kryukov G.V., Sonkin D., Reddy A., Liu M., Murray L., Berger M.F., Monahan J.E., Morais P., Meltzer J., Korejwa A., Jane-Valbuena J., Mapa F.A., Thibault J., Bric-Furlong E., Raman P., Shipway A., Engels I.H., Cheng J., Yu G.-Y.K., Yu J.-J., Aspesi P. Jr., de Silva M., Jagtap K., Jones M.D., Wang L., Hatton C., Palescandolo E., Gupta S., Mahan S., Sougnez C., Onofrio R.C., Liefeld T., MacConaill L.E., Winckler W., Reich M., Li N.-X., Mesirov J.P., Gabriel S.B., Getz G., Ardlie K., Chan V., Myer V.E., Weber B.L., Porter J., Warmuth M., Finan P., Harris J.L., Meyerson M.L., Golub T.R., Morrissey M.P., Sellers W.R., Schlegel R., Garraway L.A.
The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity.
Nature 483:603-607(2012)
PubMed=24002593; DOI=10.1038/bjc.2013.452
Wu D., Pang Y., Wilkerson M.D., Wang D., Hammerman P.S., Liu J.S.
Gene-expression data integration to squamous cell lung cancer subtypes reveals drug sensitivity.
Br. J. Cancer 109:1599-1608(2013)
PubMed=25984343; DOI=10.1038/sdata.2014.35
Cowley G.S., Weir B.A., Vazquez F., Tamayo P., Scott J.A., Rusin S., East-Seletsky A., Ali L.D., Gerath W.F.J., Pantel S.E., Lizotte P.H., Jiang G.-Z., Hsiao J., Tsherniak A., Dwinell E., Aoyama S., Okamoto M., Harrington W., Gelfand E.T., Green T.M., Tomko M.J., Gopal S., Wong T.C., Li H.-B., Howell S., Stransky N., Liefeld T., Jang D., Bistline J., Meyers B.H., Armstrong S.A., Anderson K.C., Stegmaier K., Reich M., Pellman D., Boehm J.S., Mesirov J.P., Golub T.R., Root D.E., Hahn W.C.
Parallel genome-scale loss of function screens in 216 cancer cell lines for the identification of context-specific genetic dependencies.
Sci. Data 1:140035-140035(2014)
PubMed=26589293; DOI=10.1186/s13073-015-0240-5
Scholtalbers J., Boegel S., Bukur T., Byl M., Goerges S., Sorn P., Loewer M., Sahin U., Castle J.C.
TCLP: an online cancer cell line catalogue integrating HLA type, predicted neo-epitopes, virus and gene expression.
Genome Med. 7:118.1-118.7(2015)
PubMed=27397505; DOI=10.1016/j.cell.2016.06.017
Iorio F., Knijnenburg T.A., Vis D.J., Bignell G.R., Menden M.P., Schubert M., Aben N., Goncalves E., Barthorpe S., Lightfoot H., Cokelaer T., Greninger P., van Dyk E., Chang H., de Silva H., Heyn H., Deng X.-M., Egan R.K., Liu Q.-S., Mironenko T., Mitropoulos X., Richardson L., Wang J.-H., Zhang T.-H., Moran S., Sayols S., Soleimani M., Tamborero D., Lopez-Bigas N., Ross-Macdonald P., Esteller M., Gray N.S., Haber D.A., Stratton M.R., Benes C.H., Wessels L.F.A., Saez-Rodriguez J., McDermott U., Garnett M.J.
A landscape of pharmacogenomic interactions in cancer.
Cell 166:740-754(2016)
PubMed=30894373; DOI=10.1158/0008-5472.CAN-18-2747
Dutil J., Chen Z.-H., Monteiro A.N.A., Teer J.K., Eschrich S.A.
An interactive resource to probe genetic diversity and estimated ancestry in cancer cell lines.
Cancer Res. 79:1263-1273(2019)
PubMed=30971826; DOI=10.1038/s41586-019-1103-9
Behan F.M., Iorio F., Picco G., Goncalves E., Beaver C.M., Migliardi G., Santos R., Rao Y., Sassi F., Pinnelli M., Ansari R., Harper S., Jackson D.A., McRae R., Pooley R., Wilkinson P., van der Meer D.J., Dow D., Buser-Doepner C.A., Bertotti A., Trusolino L., Stronach E.A., Saez-Rodriguez J., Yusa K., Garnett M.J.
Prioritization of cancer therapeutic targets using CRISPR-Cas9 screens.
Nature 568:511-516(2019)
PubMed=31068700; DOI=10.1038/s41586-019-1186-3
Ghandi M., Huang F.W., Jane-Valbuena J., Kryukov G.V., Lo C.C., McDonald E.R. III, Barretina J.G., Gelfand E.T., Bielski C.M., Li H.-X., Hu K., Andreev-Drakhlin A.Y., Kim J., Hess J.M., Haas B.J., Aguet F., Weir B.A., Rothberg M.V., Paolella B.R., Lawrence M.S., Akbani R., Lu Y.-L., Tiv H.L., Gokhale P.C., de Weck A., Mansour A.A., Oh C., Shih J., Hadi K., Rosen Y., Bistline J., Venkatesan K., Reddy A., Sonkin D., Liu M., Lehar J., Korn J.M., Porter D.A., Jones M.D., Golji J., Caponigro G., Taylor J.E., Dunning C.M., Creech A.L., Warren A.C., McFarland J.M., Zamanighomi M., Kauffmann A., Stransky N., Imielinski M., Maruvka Y.E., Cherniack A.D., Tsherniak A., Vazquez F., Jaffe J.D., Lane A.A., Weinstock D.M., Johannessen C.M., Morrissey M.P., Stegmeier F., Schlegel R., Hahn W.C., Getz G., Mills G.B., Boehm J.S., Golub T.R., Garraway L.A., Sellers W.R.
Next-generation characterization of the Cancer Cell Line Encyclopedia.
Nature 569:503-508(2019)
PubMed=31978347; DOI=10.1016/j.cell.2019.12.023
Nusinow D.P., Szpyt J., Ghandi M., Rose C.M., McDonald E.R. III, Kalocsay M., Jane-Valbuena J., Gelfand E.T., Schweppe D.K., Jedrychowski M.P., Golji J., Porter D.A., Rejtar T., Wang Y.K., Kryukov G.V., Stegmeier F., Erickson B.K., Garraway L.A., Sellers W.R., Gygi S.P.
Quantitative proteomics of the Cancer Cell Line Encyclopedia.
Cell 180:387-402.e16(2020)
PubMed=35839778; DOI=10.1016/j.ccell.2022.06.010
Goncalves E., Poulos R.C., Cai Z.-X., Barthorpe S., Manda S.S., Lucas N., Beck A., Bucio-Noble D., Dausmann M., Hall C., Hecker M., Koh J., Lightfoot H., Mahboob S., Mali I., Morris J., Richardson L., Seneviratne A.J., Shepherd R., Sykes E., Thomas F., Valentini S., Williams S.G., Wu Y.-X., Xavier D., MacKenzie K.L., Hains P.G., Tully B., Robinson P.J., Zhong Q., Garnett M.J., Reddel R.R.
Pan-cancer proteomic map of 949 human cell lines.
Cancer Cell 40:835-849.e8(2022)
 上海中乔新舟生物科技有限公司
上海中乔新舟生物科技有限公司