|
产品名称 |
BPH-1人前列腺增生细胞 |
|
货号 |
ZQ1076 |
|
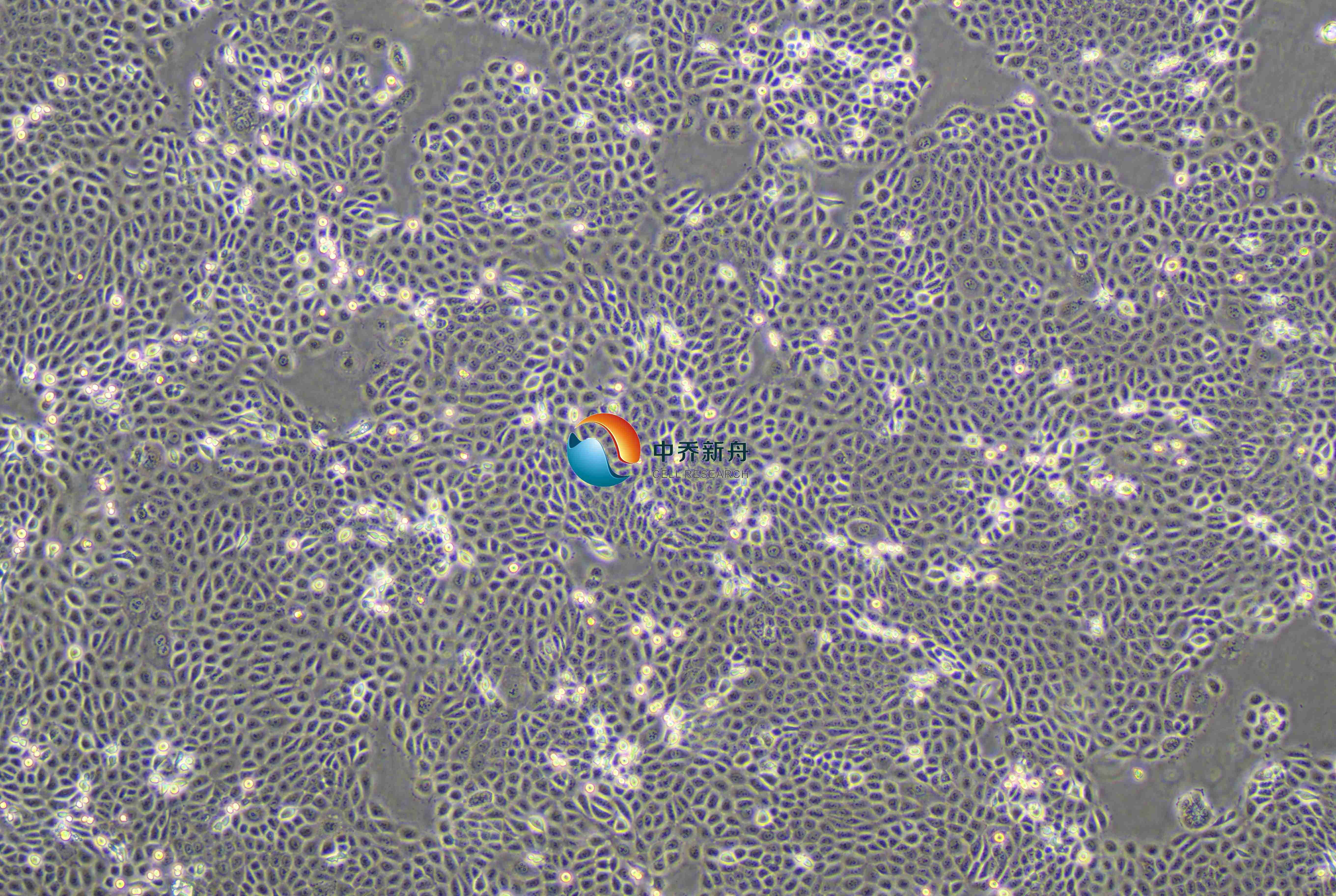
产品介绍 |
BPH-1是一种人前列腺增生细胞系,源自一位68岁男性患者的前列腺组织的上皮细胞,该细胞通过SV-40大T抗原进行永生化处理。这些细胞在前列腺生长和生理学研究中是常用的人类模型。与前列腺癌细胞系相比,BPH-1细胞表达了较高水平的雌二醇(E2)和芳香酶,这些因素被认为与前列腺增生的进展有关。这些细胞在研究前列腺增生的病因、病理生理、药物治疗效果以及前列腺疾病模型的建立中具有重要的应用价值。 |
|
种属 |
人 |
|
性别/年龄 |
男/68岁 |
|
组织 |
前列腺 |
|
疾病 |
良性前列腺增生 |
|
细胞类型 |
转化细胞系 |
|
形态学 |
上皮 |
|
生长方式 |
贴壁 |
|
倍增时间 |
~50 hours (DSMZ=ACC-143) |
|
培养基和添加剂 |
RPMI-1640(中乔新舟 货号:ZQ-200)+10%胎牛血清(中乔新舟 货号:ZQ500-A)+1%P/S(中乔新舟 货号:CSP006) |
|
推荐完全培养基货号 |
|
|
生物安全等级 |
BSL-1 |
|
培养条件 |
95%空气,5%二氧化碳;37℃ |
|
STR位点信息 |
Amelogenin: X,Y CSF1PO:10,12 D2S1338: 17 D3S1358: 16 D5S818: 11,12 D7S820: 10,11 D8S1179: 10,15 D13S317: 11,12 D16S539: 9 D18S51: 10,16 D19S433: 12,15 D21S11: 28,32.2 FGA: 21,24 (DepMap=ACH-001453; DSMZ=ACC-143; PubMed=11304728) :24 (Millipore=SCC256) Penta D :9,14 Penta E: 7,10 TH01: 6,7 TPOX: 8,12 vWA :17 |
|
抗原表达/受体表达 |
*** |
|
基因表达 |
*** |
|
保藏机构 |
DSMZ; ACC-143 ICLC; HTL11006 Millipore; SCC256 |
|
供应限制 |
仅供科研使用 |
|
货号 |
ZQ1076 |
|
发货规格 |
活细胞:T25培养瓶*1瓶或者1ml 冻存管*2支(细胞量约为1x10^6 Cells/Vial )二选一 |
|
发货形式 |
活细胞:常温运输;冻存管:干冰运输 |
|
储存温度 |
活细胞:培养箱;冻存管:液氮罐 |
|
产地 |
中国 |
|
供应限制 |
仅供科研使用 |
PubMed=7535634; DOI=10.1007/BF02631333
Hayward S.W., Dahiya R., Cunha G.R., Bartek J., Deshpande N., Narayan P.
Establishment and characterization of an immortalized but non-transformed human prostate epithelial cell line: BPH-1.
In Vitro Cell. Dev. Biol. Anim. 31:14-24(1995)
PubMed=11304728; DOI=10.1002/pros.1045
van Bokhoven A., Varella-Garcia M., Korch C.T., Hessels D., Miller G.J.
Widely used prostate carcinoma cell lines share common origins.
Prostate 47:36-51(2001)
PubMed=11719443
Phillips J.L., Hayward S.W., Wang Y.-Z., Vasselli J., Pavlovich C.P., Padilla-Nash H.M., Pezullo J.R., Ghadimi B.M., Grossfeld G.D., Rivera A., Linehan W.M., Cunha G.R., Ried T.
The consequences of chromosomal aneuploidy on gene expression profiles in a cell line model for prostate carcinogenesis.
Cancer Res. 61:8143-8149(2001)
PubMed=20164919; DOI=10.1038/nature08768
Bignell G.R., Greenman C.D., Davies H., Butler A.P., Edkins S., Andrews J.M., Buck G., Chen L., Beare D., Latimer C., Widaa S., Hinton J., Fahey C., Fu B.-Y., Swamy S., Dalgliesh G.L., Teh B.T., Deloukas P., Yang F.-T., Campbell P.J., Futreal P.A., Stratton M.R.
Signatures of mutation and selection in the cancer genome.
Nature 463:893-898(2010)
PubMed=20215515; DOI=10.1158/0008-5472.CAN-09-3458
Rothenberg S.M., Mohapatra G., Rivera M.N., Winokur D., Greninger P., Nitta M., Sadow P.M., Sooriyakumar G., Brannigan B.W., Ulman M.J., Perera R.M., Wang R., Tam A., Ma X.-J., Erlander M., Sgroi D.C., Rocco J.W., Lingen M.W., Cohen E.E.W., Louis D.N., Settleman J., Haber D.A.
A genome-wide screen for microdeletions reveals disruption of polarity complex genes in diverse human cancers.
Cancer Res. 70:2158-2164(2010)
PubMed=21248069; DOI=10.1158/0008-5472.CAN-10-1998
Li Y.-M., Alsagabi M., Fan D.-H., Bova G.S., Tewfik A.H., Dehm S.M.
Intragenic rearrangement and altered RNA splicing of the androgen receptor in a cell-based model of prostate cancer progression.
Cancer Res. 71:2108-2117(2011)
PubMed=27397505; DOI=10.1016/j.cell.2016.06.017
Iorio F., Knijnenburg T.A., Vis D.J., Bignell G.R., Menden M.P., Schubert M., Aben N., Goncalves E., Barthorpe S., Lightfoot H., Cokelaer T., Greninger P., van Dyk E., Chang H., de Silva H., Heyn H., Deng X.-M., Egan R.K., Liu Q.-S., Mironenko T., Mitropoulos X., Richardson L., Wang J.-H., Zhang T.-H., Moran S., Sayols S., Soleimani M., Tamborero D., Lopez-Bigas N., Ross-Macdonald P., Esteller M., Gray N.S., Haber D.A., Stratton M.R., Benes C.H., Wessels L.F.A., Saez-Rodriguez J., McDermott U., Garnett M.J.
A landscape of pharmacogenomic interactions in cancer.
Cell 166:740-754(2016)
PubMed=30894373; DOI=10.1158/0008-5472.CAN-18-2747
Dutil J., Chen Z.-H., Monteiro A.N.A., Teer J.K., Eschrich S.A.
An interactive resource to probe genetic diversity and estimated ancestry in cancer cell lines.
Cancer Res. 79:1263-1273(2019)
PubMed=35839778; DOI=10.1016/j.ccell.2022.06.010
Goncalves E., Poulos R.C., Cai Z.-X., Barthorpe S., Manda S.S., Lucas N., Beck A., Bucio-Noble D., Dausmann M., Hall C., Hecker M., Koh J., Lightfoot H., Mahboob S., Mali I., Morris J., Richardson L., Seneviratne A.J., Shepherd R., Sykes E., Thomas F., Valentini S., Williams S.G., Wu Y.-X., Xavier D., MacKenzie K.L., Hains P.G., Tully B., Robinson P.J., Zhong Q., Garnett M.J., Reddel R.R.
Pan-cancer proteomic map of 949 human cell lines.
Cancer Cell 40:835-849.e8(2022)
 上海中乔新舟生物科技有限公司
上海中乔新舟生物科技有限公司