|
产品名称 |
LS513 人结直肠癌细胞 |
|
货号 |
ZQ1080 |
|
产品介绍 |
LS513是一个结直肠癌细胞株,1985年从一位诊断为Dukes' C粘液分泌盲肠肿瘤的63岁白人男性患者的Bauhin阀肿瘤原位活检中建株。LS513 细胞具有粘附性,并表现出多药耐药性 (MDR),使其成为研究结肠直肠癌耐药机制的宝贵模型。这些细胞在甲基纤维素中表现出 30% 的菌落形成效率,并且在裸鼠中具有致瘤性,进一步验证了它们在致癌研究中的用途。
注意事项: |
|
种属 |
人 |
|
性别/年龄 |
男/63岁 |
|
组织 |
盲肠 |
|
疾病 |
结直肠癌 |
|
细胞类型 |
肿瘤细胞 |
|
形态学 |
上皮 |
|
生长方式 |
贴壁 |
|
倍增时间 |
33.93 hours (PubMed=25944804); 20 hours (PubMed=25984343) |
|
培养基和添加剂 |
RPMI-1640(品牌:中乔新舟 货号:ZQ-200)+10%胎牛血清(中乔新舟 货号:ZQ500-A)+1%P/S(中乔新舟 货号:CSP006) |
|
推荐完全培养基货号 |
|
|
生物安全等级 |
BSL-1 |
|
培养条件 |
95%空气,5%二氧化碳;37℃ |
|
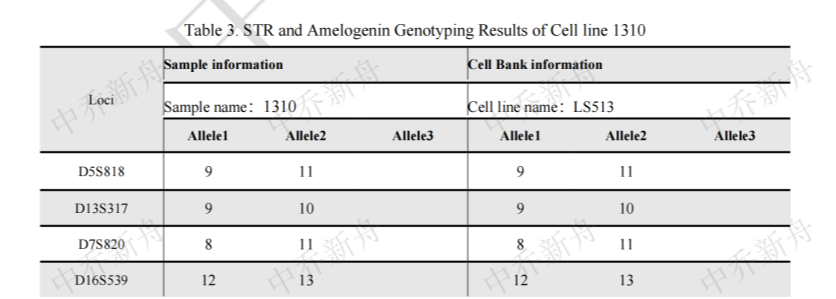
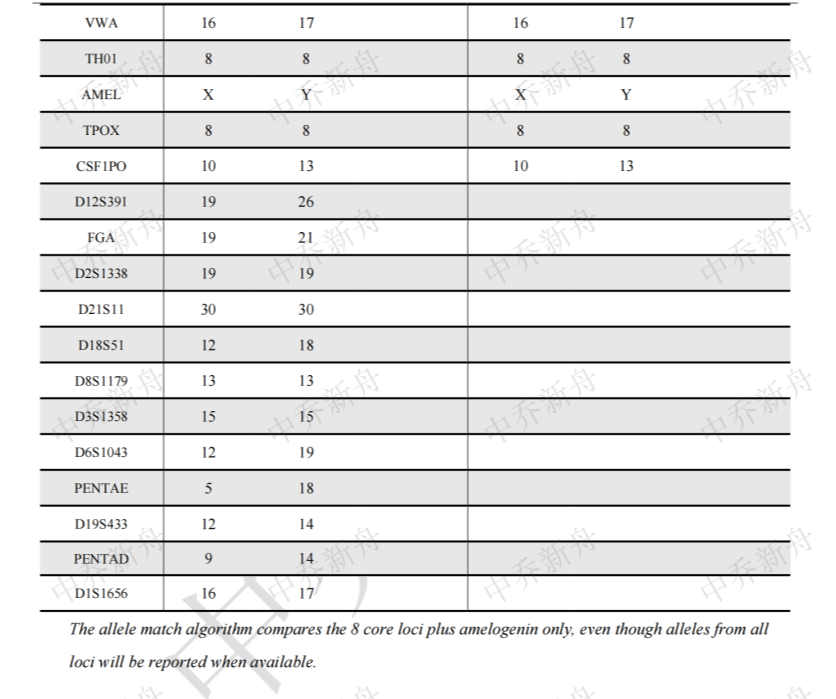
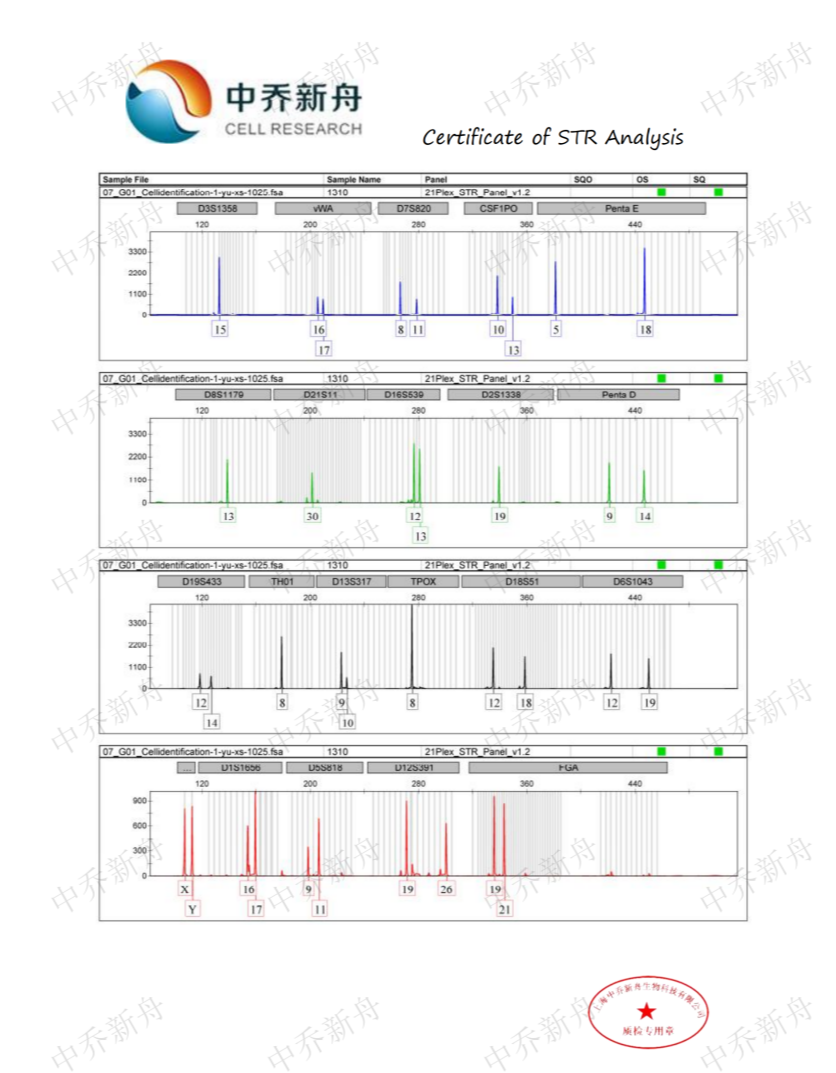
STR位点信息 |
Amelogenin: X,Y CSF1PO :10 (CLS=300457) :10,13 (ATCC=CRL-2134; Cosmic-CLP=907795; KCLB=22134; PubMed=25926053) D2S1338 :19 D3S1358 :15 D5S818: 9,11 (ATCC=CRL-2134; Cosmic-CLP=907795; KCLB=22134; PubMed=25926053) :11 (CLS=300457) D7S820: 8,11 D8S1179: 13 D13S317: 9,10 D16S539: 12,13 D18S51: 12,18 D19S433: 12,14 D21S11: 30 FGA: 19,21 Penta D: 9,14 Penta E: 5,18 TH01: 8 TPOX :8 vWA: 16,17 |
|
抗原表达/受体表达 |
*** |
|
基因表达 |
*** |
|
保藏机构 |
CLS; 300457 ATCC;CRL-2134 |
|
供应限制 |
仅供科研使用 |
|
货号 |
ZQ1080 |
|
发货规格 |
活细胞:T25培养瓶*1瓶或者1ml 冻存管*2支(细胞量约为1x10^6 Cells/Vial )二选一 |
|
发货形式 |
活细胞:常温运输;冻存管:干冰运输 |
|
储存温度 |
活细胞:培养箱;冻存管:液氮罐 |
|
产地 |
中国 |
|
供应限制 |
仅供科研使用 |
PubMed=1389533; DOI=10.1016/0959-8049(92)90031-V
Lahm H., Petral-Malec D., Yilmaz-Ceyhan A., Fischer J.R., Lorenzoni M., Givel J.-C., Odartchenko N.
Growth stimulation of a human colorectal carcinoma cell line by interleukin-1 and -6 and antagonistic effects of transforming growth factor beta 1.
Eur. J. Cancer 28:1894-1899(1992)
PubMed=1617643
Suardet L., Gaide A.-C., Calmes J.-M., Sordat B., Givel J.-C., Eliason J.F., Odartchenko N.
Responsiveness of three newly established human colorectal cancer cell lines to transforming growth factors beta 1 and beta 2.
Cancer Res. 52:3705-3712(1992)
PubMed=8895552; DOI=10.1002/(SICI)1097-0215(19960927)68:1<126::AID-IJC22>3.0.CO;2-8
Suardet L., Li C., Little J.B.
Radio-induced modulation of transforming growth factor beta1 sensitivity in a p53 wild-type human colorectal-cancer cell line.
Int. J. Cancer 68:126-131(1996)
PubMed=9000147
Cottu P.-H., Muzeau F., Estreicher A., Flejou J.-F., Iggo R.D., Thomas G., Hamelin R.
Inverse correlation between RER+ status and p53 mutation in colorectal cancer cell lines.
Oncogene 13:2727-2730(1996)
PubMed=9000572
Hoang J.-M., Cottu P.-H., Thuille B., Salmon R.J., Thomas G., Hamelin R.
BAT-26, an indicator of the replication error phenotype in colorectal cancers and cell lines.
Cancer Res. 57:300-303(1997)
PubMed=11414198; DOI=10.1007/s004320000207
Lahm H., Andre S., Hoeflich A., Fischer J.R., Sordat B., Kaltner H., Wolf E., Gabius H.-J.
Comprehensive galectin fingerprinting in a panel of 61 human tumor cell lines by RT-PCR and its implications for diagnostic and therapeutic procedures.
J. Cancer Res. Clin. Oncol. 127:375-386(2001)
PubMed=11526487; DOI=10.1038/sj.onc.1204611
Gayet J., Zhou X.-P., Duval A., Rolland S., Hoang J.-M., Cottu P.-H., Hamelin R.
Extensive characterization of genetic alterations in a series of human colorectal cancer cell lines.
Oncogene 20:5025-5032(2001)
PubMed=12068308; DOI=10.1038/nature00766
Davies H., Bignell G.R., Cox C., Stephens P.J., Edkins S., Clegg S., Teague J.W., Woffendin H., Garnett M.J., Bottomley W., Davis N., Dicks E., Ewing R., Floyd Y., Gray K., Hall S., Hawes R., Hughes J., Kosmidou V., Menzies A., Mould C., Parker A., Stevens C., Watt S., Hooper S., Wilson R., Jayatilake H., Gusterson B.A., Cooper C.S., Shipley J.M., Hargrave D., Pritchard-Jones K., Maitland N.J., Chenevix-Trench G., Riggins G.J., Bigner D.D., Palmieri G., Cossu A., Flanagan A.M., Nicholson A., Ho J.W.C., Leung S.Y., Yuen S.T., Weber B.L., Seigler H.F., Darrow T.L., Paterson H.F., Marais R., Marshall C.J., Wooster R., Stratton M.R., Futreal P.A.
Mutations of the BRAF gene in human cancer.
Nature 417:949-954(2002)
PubMed=16418264; DOI=10.1073/pnas.0510146103
Liu Y., Bodmer W.F.
Analysis of P53 mutations and their expression in 56 colorectal cancer cell lines.
Proc. Natl. Acad. Sci. U.S.A. 103:976-981(2006)
PubMed=16854228; DOI=10.1186/1476-4598-5-29
Bandres Elizalde E.M., Cubedo E., Agirre X., Malumbres R., Zarate R., Ramirez N., Abajo A., Navarro A., Moreno I., Monzo M., Garcia-Foncillas J.
Identification by real-time PCR of 13 mature microRNAs differentially expressed in colorectal cancer and non-tumoral tissues.
Mol. Cancer 5:29.1-29.10(2006)
PubMed=18258742; DOI=10.1073/pnas.0712176105
Emaduddin M., Bicknell D.C., Bodmer W.F., Feller S.M.
Cell growth, global phosphotyrosine elevation, and c-Met phosphorylation through Src family kinases in colorectal cancer cells.
Proc. Natl. Acad. Sci. U.S.A. 105:2358-2362(2008)
PubMed=20164919; DOI=10.1038/nature08768
Bignell G.R., Greenman C.D., Davies H., Butler A.P., Edkins S., Andrews J.M., Buck G., Chen L., Beare D., Latimer C., Widaa S., Hinton J., Fahey C., Fu B.-Y., Swamy S., Dalgliesh G.L., Teh B.T., Deloukas P., Yang F.-T., Campbell P.J., Futreal P.A., Stratton M.R.
Signatures of mutation and selection in the cancer genome.
Nature 463:893-898(2010)
PubMed=20570890; DOI=10.1158/0008-5472.CAN-10-0192
Janakiraman M., Vakiani E., Zeng Z.-S., Pratilas C.A., Taylor B.S., Chitale D., Halilovic E., Wilson M., Huberman K., Ricarte Filho J.C.M., Persaud Y., Levine D.A., Fagin J.A., Jhanwar S.C., Mariadason J.M., Lash A., Ladanyi M., Saltz L.B., Heguy A., Paty P.B., Solit D.B.
Genomic and biological characterization of exon 4 KRAS mutations in human cancer.
Cancer Res. 70:5901-5911(2010)
PubMed=20606684; DOI=10.1038/sj.bjc.6605780
Bracht K., Nicholls A.M., Liu Y., Bodmer W.F.
5-fluorouracil response in a large panel of colorectal cancer cell lines is associated with mismatch repair deficiency.
Br. J. Cancer 103:340-346(2010)
PubMed=22460905; DOI=10.1038/nature11003
Barretina J.G., Caponigro G., Stransky N., Venkatesan K., Margolin A.A., Kim S., Wilson C.J., Lehar J., Kryukov G.V., Sonkin D., Reddy A., Liu M., Murray L., Berger M.F., Monahan J.E., Morais P., Meltzer J., Korejwa A., Jane-Valbuena J., Mapa F.A., Thibault J., Bric-Furlong E., Raman P., Shipway A., Engels I.H., Cheng J., Yu G.-Y.K., Yu J.-J., Aspesi P. Jr., de Silva M., Jagtap K., Jones M.D., Wang L., Hatton C., Palescandolo E., Gupta S., Mahan S., Sougnez C., Onofrio R.C., Liefeld T., MacConaill L.E., Winckler W., Reich M., Li N.-X., Mesirov J.P., Gabriel S.B., Getz G., Ardlie K., Chan V., Myer V.E., Weber B.L., Porter J., Warmuth M., Finan P., Harris J.L., Meyerson M.L., Golub T.R., Morrissey M.P., Sellers W.R., Schlegel R., Garraway L.A.
The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity.
Nature 483:603-607(2012)
PubMed=23272949; DOI=10.1186/1755-8794-5-66
Schlicker A., Beran G., Chresta C.M., McWalter G., Pritchard A., Weston S., Runswick S., Davenport S., Heathcote K., Castro D.A., Orphanides G., French T., Wessels L.F.A.
Subtypes of primary colorectal tumors correlate with response to targeted treatment in colorectal cell lines.
BMC Med. Genomics 5:66.1-66.15(2012)
PubMed=23932154; DOI=10.1016/j.radonc.2013.06.032
Salendo J., Spitzner M., Kramer F., Zhang X., Jo P., Wolff H.A., Kitz J., Kaulfuss S., Beissbarth T., Dobbelstein M., Ghadimi M., Grade M., Gaedcke J.
Identification of a microRNA expression signature for chemoradiosensitivity of colorectal cancer cells, involving miRNAs-320a, -224, -132 and let7g.
Radiother. Oncol. 108:451-457(2013)
PubMed=24755471; DOI=10.1158/0008-5472.CAN-14-0013
Mouradov D., Sloggett C., Jorissen R.N., Love C.G., Li S., Burgess A.W., Arango D., Strausberg R.L., Buchanan D., Wormald S., O'Connor L., Wilding J.L., Bicknell D.C., Tomlinson I.P.M., Bodmer W.F., Mariadason J.M., Sieber O.M.
Colorectal cancer cell lines are representative models of the main molecular subtypes of primary cancer.
Cancer Res. 74:3238-3247(2014)
PubMed=25984343; DOI=10.1038/sdata.2014.35
Cowley G.S., Weir B.A., Vazquez F., Tamayo P., Scott J.A., Rusin S., East-Seletsky A., Ali L.D., Gerath W.F.J., Pantel S.E., Lizotte P.H., Jiang G.-Z., Hsiao J., Tsherniak A., Dwinell E., Aoyama S., Okamoto M., Harrington W., Gelfand E.T., Green T.M., Tomko M.J., Gopal S., Wong T.C., Li H.-B., Howell S., Stransky N., Liefeld T., Jang D., Bistline J., Meyers B.H., Armstrong S.A., Anderson K.C., Stegmaier K., Reich M., Pellman D., Boehm J.S., Mesirov J.P., Golub T.R., Root D.E., Hahn W.C.
Parallel genome-scale loss of function screens in 216 cancer cell lines for the identification of context-specific genetic dependencies.
Sci. Data 1:140035-140035(2014)
PubMed=25926053; DOI=10.1038/ncomms8002
Medico E., Russo M., Picco G., Cancelliere C., Valtorta E., Corti G., Buscarino M., Isella C., Lamba S., Martinoglio B., Veronese S., Siena S., Sartore-Bianchi A., Beccuti M., Mottolese M., Linnebacher M., Cordero F., Di Nicolantonio F., Bardelli A.
The molecular landscape of colorectal cancer cell lines unveils clinically actionable kinase targets.
Nat. Commun. 6:7002.1-7002.10(2015)
PubMed=25944804; DOI=10.1158/1078-0432.CCR-14-2457
Bazzocco S., Dopeso H., Carton-Garcia F., Macaya I., Andretta E., Chionh F., Rodrigues P., Garrido M., Alazzouzi H., Nieto R., Sanchez A., Schwartz S. Jr., Bilic J., Mariadason J.M., Arango D.
Highly expressed genes in rapidly proliferating tumor cells as new targets for colorectal cancer treatment.
Clin. Cancer Res. 21:3695-3704(2015)
PubMed=27397505; DOI=10.1016/j.cell.2016.06.017
Iorio F., Knijnenburg T.A., Vis D.J., Bignell G.R., Menden M.P., Schubert M., Aben N., Goncalves E., Barthorpe S., Lightfoot H., Cokelaer T., Greninger P., van Dyk E., Chang H., de Silva H., Heyn H., Deng X.-M., Egan R.K., Liu Q.-S., Mironenko T., Mitropoulos X., Richardson L., Wang J.-H., Zhang T.-H., Moran S., Sayols S., Soleimani M., Tamborero D., Lopez-Bigas N., Ross-Macdonald P., Esteller M., Gray N.S., Haber D.A., Stratton M.R., Benes C.H., Wessels L.F.A., Saez-Rodriguez J., McDermott U., Garnett M.J.
A landscape of pharmacogenomic interactions in cancer.
Cell 166:740-754(2016)
PubMed=28854368; DOI=10.1016/j.celrep.2017.08.010
Roumeliotis T.I., Williams S.P., Goncalves E., Alsinet C., Del Castillo Velasco-Herrera M., Aben N., Ghavidel F.Z., Michaut M., Schubert M., Price S., Wright J.C., Yu L., Yang M., Dienstmann R., Guinney J., Beltrao P., Brazma A., Pardo M., Stegle O., Adams D.J., Wessels L.F.A., Saez-Rodriguez J., McDermott U., Choudhary J.S.
Genomic determinants of protein abundance variation in colorectal cancer cells.
Cell Rep. 20:2201-2214(2017)
PubMed=29101300; DOI=10.15252/msb.20177701
Frejno M., Zenezini Chiozzi R., Wilhelm M., Koch H., Zheng R.-S., Klaeger S., Ruprecht B., Meng C., Kramer K., Jarzab A., Heinzlmeir S., Johnstone E., Domingo E., Kerr D., Jesinghaus M., Slotta-Huspenina J., Weichert W., Knapp S., Feller S.M., Kuster B.
Pharmacoproteomic characterisation of human colon and rectal cancer.
Mol. Syst. Biol. 13:951-951(2017)
PubMed=29444439; DOI=10.1016/j.celrep.2018.01.051
Yuan T.L., Amzallag A., Bagni R., Yi M., Afghani S., Burgan W., Fer N., Strathern L.A., Powell K., Smith B., Waters A.M., Drubin D.A., Thomson T., Liao R., Greninger P., Stein G.T., Murchie E., Cortez E., Egan R.K., Procter L., Bess M., Cheng K.T., Lee C.-S., Lee L.C., Fellmann C., Stephens R., Luo J., Lowe S.W., Benes C.H., McCormick F.
Differential effector engagement by oncogenic KRAS.
Cell Rep. 22:1889-1902(2018)
PubMed=30894373; DOI=10.1158/0008-5472.CAN-18-2747
Dutil J., Chen Z.-H., Monteiro A.N.A., Teer J.K., Eschrich S.A.
An interactive resource to probe genetic diversity and estimated ancestry in cancer cell lines.
Cancer Res. 79:1263-1273(2019)
PubMed=30971826; DOI=10.1038/s41586-019-1103-9
Behan F.M., Iorio F., Picco G., Goncalves E., Beaver C.M., Migliardi G., Santos R., Rao Y., Sassi F., Pinnelli M., Ansari R., Harper S., Jackson D.A., McRae R., Pooley R., Wilkinson P., van der Meer D.J., Dow D., Buser-Doepner C.A., Bertotti A., Trusolino L., Stronach E.A., Saez-Rodriguez J., Yusa K., Garnett M.J.
Prioritization of cancer therapeutic targets using CRISPR-Cas9 screens.
Nature 568:511-516(2019)
PubMed=31068700; DOI=10.1038/s41586-019-1186-3
Ghandi M., Huang F.W., Jane-Valbuena J., Kryukov G.V., Lo C.C., McDonald E.R. III, Barretina J.G., Gelfand E.T., Bielski C.M., Li H.-X., Hu K., Andreev-Drakhlin A.Y., Kim J., Hess J.M., Haas B.J., Aguet F., Weir B.A., Rothberg M.V., Paolella B.R., Lawrence M.S., Akbani R., Lu Y.-L., Tiv H.L., Gokhale P.C., de Weck A., Mansour A.A., Oh C., Shih J., Hadi K., Rosen Y., Bistline J., Venkatesan K., Reddy A., Sonkin D., Liu M., Lehar J., Korn J.M., Porter D.A., Jones M.D., Golji J., Caponigro G., Taylor J.E., Dunning C.M., Creech A.L., Warren A.C., McFarland J.M., Zamanighomi M., Kauffmann A., Stransky N., Imielinski M., Maruvka Y.E., Cherniack A.D., Tsherniak A., Vazquez F., Jaffe J.D., Lane A.A., Weinstock D.M., Johannessen C.M., Morrissey M.P., Stegmeier F., Schlegel R., Hahn W.C., Getz G., Mills G.B., Boehm J.S., Golub T.R., Garraway L.A., Sellers W.R.
Next-generation characterization of the Cancer Cell Line Encyclopedia.
Nature 569:503-508(2019)
PubMed=31978347; DOI=10.1016/j.cell.2019.12.023
Nusinow D.P., Szpyt J., Ghandi M., Rose C.M., McDonald E.R. III, Kalocsay M., Jane-Valbuena J., Gelfand E.T., Schweppe D.K., Jedrychowski M.P., Golji J., Porter D.A., Rejtar T., Wang Y.K., Kryukov G.V., Stegmeier F., Erickson B.K., Garraway L.A., Sellers W.R., Gygi S.P.
Quantitative proteomics of the Cancer Cell Line Encyclopedia.
Cell 180:387-402.e16(2020)
PubMed=35839778; DOI=10.1016/j.ccell.2022.06.010
Goncalves E., Poulos R.C., Cai Z.-X., Barthorpe S., Manda S.S., Lucas N., Beck A., Bucio-Noble D., Dausmann M., Hall C., Hecker M., Koh J., Lightfoot H., Mahboob S., Mali I., Morris J., Richardson L., Seneviratne A.J., Shepherd R., Sykes E., Thomas F., Valentini S., Williams S.G., Wu Y.-X., Xavier D., MacKenzie K.L., Hains P.G., Tully B., Robinson P.J., Zhong Q., Garnett M.J., Reddel R.R.
Pan-cancer proteomic map of 949 human cell lines.
Cancer Cell 40:835-849.e8(2022)
 上海中乔新舟生物科技有限公司
上海中乔新舟生物科技有限公司