|
产品名称 |
HGC-27人胃癌细胞(未分化) |
|
货号 |
ZQ0192 |
|
产品介绍 |
HGC-27是一种人胃癌细胞系,源自一名成年胃癌患者的淋巴结转移灶,这种细胞系由未分化胃癌细胞组成,能够产生并分泌黏液素。该细胞系表现出上皮形态。
HGC-27细胞系在胃癌研究中的应用有以下案例:黄芩素对胃癌HGC-27细胞上皮间充质转化进程的影响研究、紫杉醇耐药人胃癌细胞HGC-27/PTX的建立及其特征鉴定、虫草素对人胃癌细胞系HGC-27的影响及分子机制、过表达FAF1对胃癌细胞HGC-27增殖及凋亡的影响、SOX9在胃癌组织中的表达及其对胃癌细胞HGC-27生物学功能的影响、G蛋白偶联受体34敲低损害HGC-27胃癌细胞的体外增殖和迁移、人胃癌HGC-27细胞及其肿瘤球细胞裸小鼠皮下移植的研究等。 |
|
种属 |
人 |
|
性别/年龄 |
***/成年人 |
|
组织 |
胃,淋巴结转移 |
|
疾病 |
胃癌 |
|
细胞类型 |
肿瘤细胞 |
|
形态学 |
上皮细胞 |
|
生长方式 |
贴壁 |
|
倍增时间 |
大约17小时 |
|
培养基和添加剂 |
RPMI-1640(中乔新舟 货号:ZQ-200)+20%胎牛血清(中乔新舟 货号:ZQ0500)+1%双抗(中乔新舟 货号:CSP006) |
|
推荐完全培养基货号 |
|
|
生物安全等级 |
BSL-1 |
|
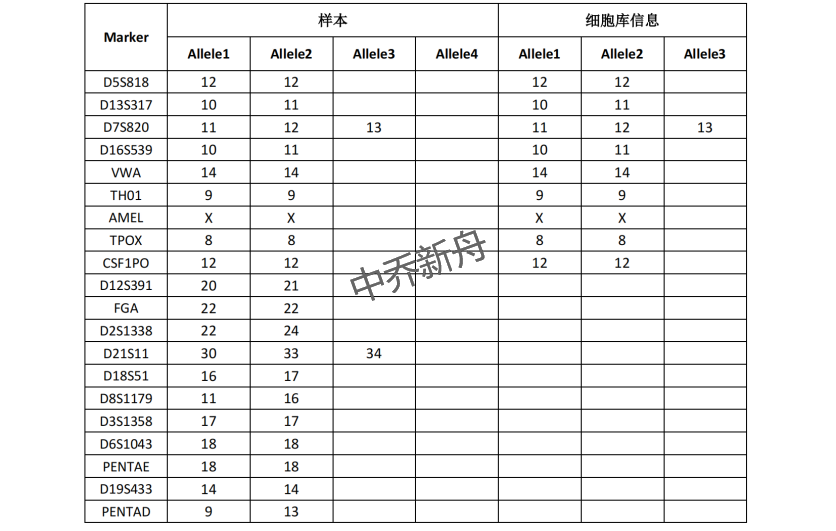
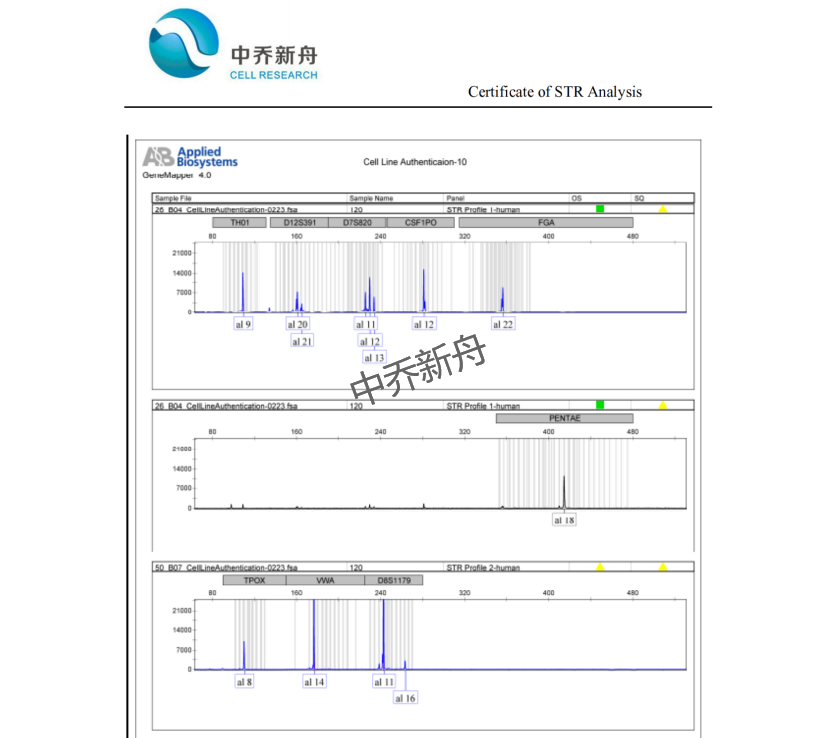
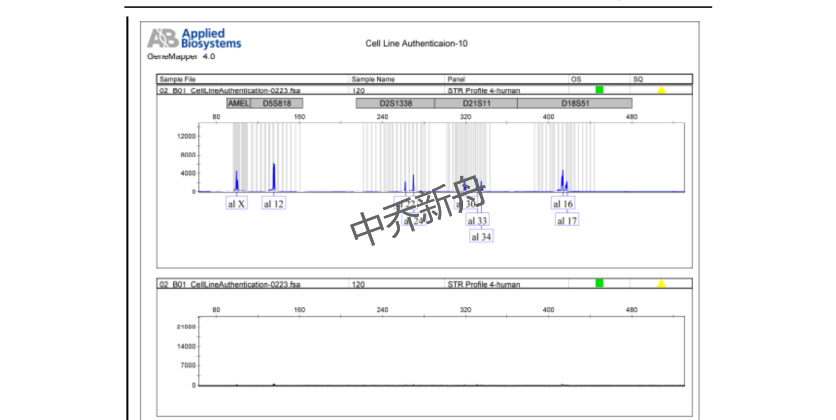
STR位点信息 |
Amelogenin: X |
|
培养条件 |
95%空气,5%二氧化碳;37℃ |
|
抗原表达/受体表达 |
*** |
|
基因表达 |
*** |
|
保藏机构 |
BCRJ; 0310 ;ECACC; 94042256 ;ICLC; HTL95028 ;KCB; KCB 201601YJ ;RCB; RCB0500 |
|
供应限制 |
仅供科研使用 |
|
货号 |
ZM0192 |
|
发货规格 |
活细胞:T25培养瓶*1瓶或者1ml 冻存管*2支(细胞量约为1x10^6 Cells/Vial)二选一 |
|
发货形式 |
活细胞:常温运输;冻存管:干冰运输 |
|
储存温度 |
活细胞:培养箱;冻存管:液氮罐 |
|
产地 |
中国 |
|
供应限制 |
仅供科研使用 |

| 货号 | 产品名称 | 规格 | 价格 | 指令 |
 上海中乔新舟生物科技有限公司
上海中乔新舟生物科技有限公司