|
产品名称 |
VCaP人前列腺癌细胞 |
|
货号 |
ZQ0353 |
|
产品介绍 |
VCaP(前列腺Vertebral-Cancer)细胞株是研究前列腺癌的重要模型,源于人类前列腺癌的脊椎转移,它在体外和体内均对雄激素敏感。它的建立是为了提供一个相关的体外模型,用于研究前列腺癌的生物学及其转移过程,特别是关注该疾病的激素难治性阶段。VCaP细胞以高水平表达前列腺特异性抗原(PSA)和雄激素受体(AR)而闻名,这使得它们与雄激素受体信号通路和抗雄激素治疗耐药机制的研究高度相关。
特别注意: |
|
种属 |
人 |
|
性别/年龄 |
男/59岁 |
|
组织 |
前列腺;来源于转移部位:脊椎转移 |
|
疾病 |
癌症 |
|
细胞类型 |
上皮细胞 |
|
形态学 |
上皮的 |
|
生长方式 |
贴壁 |
|
倍增时间 |
大约51~220小时 |
|
培养基和添加剂 |
DMEM高糖(品牌:中乔新舟 货号:ZQ-100)+10%胎牛血清(中乔新舟 货号:ZQ0500)+1%P/S(中乔新舟 货号:CSP006) |
|
推荐完全培养基货号 |
|
|
生物安全等级 |
BSL-2 |
|
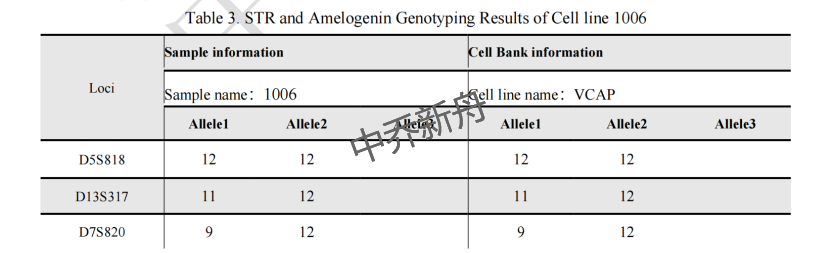
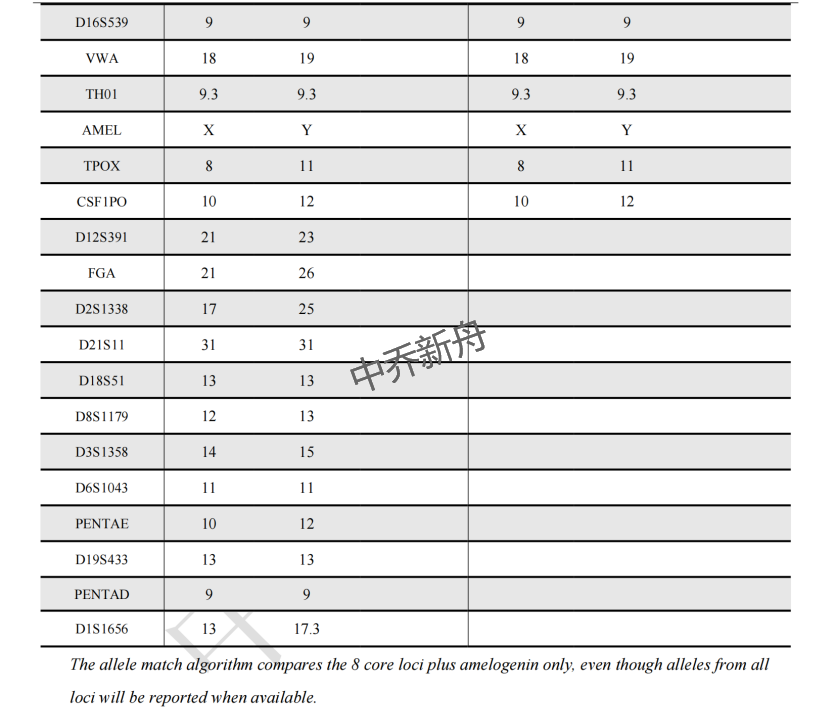
STR位点信息 |
Amelogenin: X,Y CSF1PO: 10,12 D13S317: 11,12 D16S539: 9 D5S818: 12 D7S820: 9,12 TH01: 9.3 TPOX: 8,11 vWA: 18,19 |
|
培养条件 |
95%空气,5%二氧化碳;37℃ |
|
抗原表达/受体表达 |
*** |
|
基因表达 |
*** |
|
保藏机构 |
ATCC; CRL-2876 ECACC; 06020201 |
|
供应限制 |
仅供科研使用 |
|
货号 |
ZQ0353 |
|
发货规格 |
活细胞:T25培养瓶*1瓶或者1ml 冻存管*2支(细胞量约为1x10^6 Cells/Vial)二选一 |
|
发货形式 |
活细胞:常温运输;冻存管:干冰运输 |
|
储存温度 |
活细胞:培养箱;冻存管:液氮罐 |
|
产地 |
中国 |
|
供应限制 |
仅供科研使用 |

| 货号 | 产品名称 | 规格 | 价格 | 指令 |
 上海中乔新舟生物科技有限公司
上海中乔新舟生物科技有限公司