|
产品名称 |
VCaP人前列腺癌细胞 |
|
货号 |
ZQ0353 |
|
产品介绍 |
VCaP(前列腺Vertebral-Cancer)细胞株是研究前列腺癌的重要模型,源于人类前列腺癌的脊椎转移,它在体外和体内均对雄激素敏感。它的建立是为了提供一个相关的体外模型,用于研究前列腺癌的生物学及其转移过程,特别是关注该疾病的激素难治性阶段。VCaP细胞以高水平表达前列腺特异性抗原(PSA)和雄激素受体(AR)而闻名,这使得它们与雄激素受体信号通路和抗雄激素治疗耐药机制的研究高度相关。
特别注意: |
|
种属 |
人 |
|
性别/年龄 |
男/59岁 |
|
组织 |
前列腺;来源于转移部位:脊椎转移 |
|
疾病 |
癌症 |
|
细胞类型 |
上皮细胞 |
|
形态学 |
上皮的 |
|
生长方式 |
贴壁 |
|
倍增时间 |
大约51~220小时 |
|
培养基和添加剂 |
DMEM高糖(品牌:中乔新舟 货号:ZQ-100)+10%胎牛血清(中乔新舟 货号:ZQ0500)+1%P/S(中乔新舟 货号:CSP006) |
|
推荐完全培养基货号 |
|
|
生物安全等级 |
BSL-2 |
|
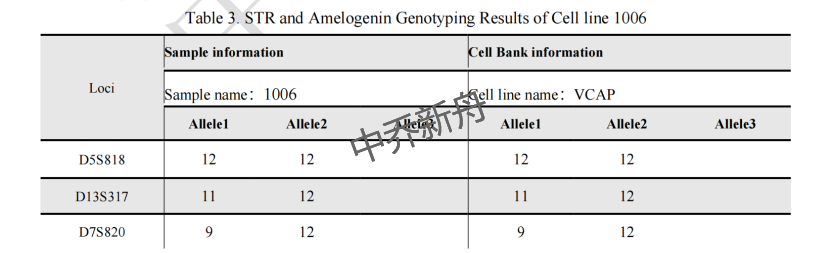
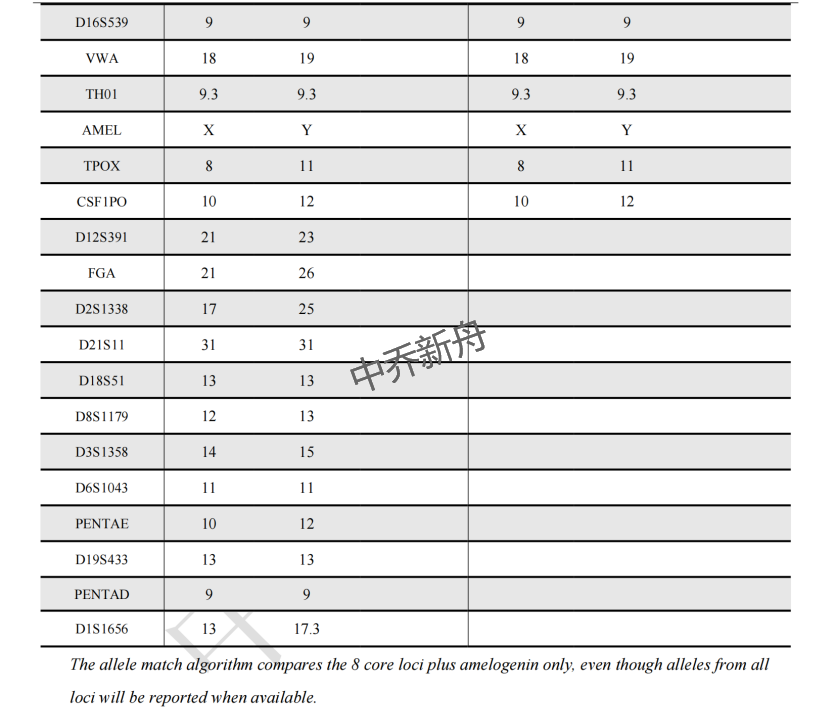
STR位点信息 |
Amelogenin: X,Y CSF1PO: 10,12 D13S317: 11,12 D16S539: 9 D5S818: 12 D7S820: 9,12 TH01: 9.3 TPOX: 8,11 vWA: 18,19 |
|
培养条件 |
95%空气,5%二氧化碳;37℃ |
|
抗原表达/受体表达 |
*** |
|
基因表达 |
*** |
|
保藏机构 |
ATCC; CRL-2876 ECACC; 06020201 |
|
供应限制 |
仅供科研使用 |
|
货号 |
ZQ0353 |
|
发货规格 |
活细胞:T25培养瓶*1瓶或者1ml 冻存管*2支(细胞量约为1x10^6 Cells/Vial)二选一 |
|
发货形式 |
活细胞:常温运输;冻存管:干冰运输 |
|
储存温度 |
活细胞:培养箱;冻存管:液氮罐 |
|
产地 |
中国 |
|
供应限制 |
仅供科研使用 |
PubMed=11317522
Korenchuk S., Lehr J.E., McLean L., Lee Y.-G., Whitney S., Vessella R.L., Lin D.L., Pienta K.J.
VCaP, a cell-based model system of human prostate cancer.
In Vivo 15:163-168(2001)
PubMed=14518029; DOI=10.1002/pros.10290
van Bokhoven A., Varella-Garcia M., Korch C.T., Johannes W.U., Smith E.E., Miller H.L., Nordeen S.K., Miller G.J., Lucia M.S.
Molecular characterization of human prostate carcinoma cell lines.
Prostate 57:205-225(2003)
PubMed=14518030; DOI=10.1002/pros.10291
van Bokhoven A., Caires A., Di Maria M., Schulte A.P., Lucia M.S., Nordeen S.K., Miller G.J., Varella-Garcia M.
Spectral karyotype (SKY) analysis of human prostate carcinoma cell lines.
Prostate 57:226-244(2003)
PubMed=21698104; DOI=10.1371/journal.pone.0020874
Sfanos K.S., Aloia A.L., Hicks J.L., Esopi D.M., Steranka J.P., Shao W., Sanchez-Martinez S., Yegnasubramanian S., Burns K.H., Rein A., De Marzo A.M.
Identification of replication competent murine gammaretroviruses in commonly used prostate cancer cell lines.
PLoS ONE 6:E20874-E20874(2011)
PubMed=22460905; DOI=10.1038/nature11003
Barretina J.G., Caponigro G., Stransky N., Venkatesan K., Margolin A.A., Kim S., Wilson C.J., Lehar J., Kryukov G.V., Sonkin D., Reddy A., Liu M., Murray L., Berger M.F., Monahan J.E., Morais P., Meltzer J., Korejwa A., Jane-Valbuena J., Mapa F.A., Thibault J., Bric-Furlong E., Raman P., Shipway A., Engels I.H., Cheng J., Yu G.-Y.K., Yu J.-J., Aspesi P. Jr., de Silva M., Jagtap K., Jones M.D., Wang L., Hatton C., Palescandolo E., Gupta S., Mahan S., Sougnez C., Onofrio R.C., Liefeld T., MacConaill L.E., Winckler W., Reich M., Li N.-X., Mesirov J.P., Gabriel S.B., Getz G., Ardlie K., Chan V., Myer V.E., Weber B.L., Porter J., Warmuth M., Finan P., Harris J.L., Meyerson M.L., Golub T.R., Morrissey M.P., Sellers W.R., Schlegel R., Garraway L.A.
The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity.
Nature 483:603-607(2012)
PubMed=23447416; DOI=10.1002/path.4186
Weier C., Haffner M.C., Mosbruger T.L., Esopi D.M., Hicks J.L., Zheng Q.-Z., Fedor H., Isaacs W.B., De Marzo A.M., Nelson W.G., Yegnasubramanian S.
Nucleotide resolution analysis of TMPRSS2 and ERG rearrangements in prostate cancer.
J. Pathol. 230:174-183(2013)
PubMed=23615946; DOI=10.1007/s00439-013-1308-1
Teles Alves I., Hiltemann S., Hartjes T., van der Spek P.J., Stubbs A., Trapman J., Jenster G.
Gene fusions by chromothripsis of chromosome 5q in the VCaP prostate cancer cell line.
Hum. Genet. 132:709-713(2013)
PubMed=23671654; DOI=10.1371/journal.pone.0063056
Lu Y.-H., Soong T.D., Elemento O.
A novel approach for characterizing microsatellite instability in cancer cells.
PLoS ONE 8:E63056-E63056(2013)
PubMed=25984343; DOI=10.1038/sdata.2014.35
Cowley G.S., Weir B.A., Vazquez F., Tamayo P., Scott J.A., Rusin S., East-Seletsky A., Ali L.D., Gerath W.F.J., Pantel S.E., Lizotte P.H., Jiang G.-Z., Hsiao J., Tsherniak A., Dwinell E., Aoyama S., Okamoto M., Harrington W., Gelfand E.T., Green T.M., Tomko M.J., Gopal S., Wong T.C., Li H.-B., Howell S., Stransky N., Liefeld T., Jang D., Bistline J., Meyers B.H., Armstrong S.A., Anderson K.C., Stegmaier K., Reich M., Pellman D., Boehm J.S., Mesirov J.P., Golub T.R., Root D.E., Hahn W.C.
Parallel genome-scale loss of function screens in 216 cancer cell lines for the identification of context-specific genetic dependencies.
Sci. Data 1:140035-140035(2014)
PubMed=25877200; DOI=10.1038/nature14397
Yu M., Selvaraj S.K., Liang-Chu M.M.Y., Aghajani S., Busse M., Yuan J., Lee G., Peale F.V., Klijn C., Bourgon R., Kaminker J.S., Neve R.M.
A resource for cell line authentication, annotation and quality control.
Nature 520:307-311(2015)
PubMed=27397505; DOI=10.1016/j.cell.2016.06.017
Iorio F., Knijnenburg T.A., Vis D.J., Bignell G.R., Menden M.P., Schubert M., Aben N., Goncalves E., Barthorpe S., Lightfoot H., Cokelaer T., Greninger P., van Dyk E., Chang H., de Silva H., Heyn H., Deng X.-M., Egan R.K., Liu Q.-S., Mironenko T., Mitropoulos X., Richardson L., Wang J.-H., Zhang T.-H., Moran S., Sayols S., Soleimani M., Tamborero D., Lopez-Bigas N., Ross-Macdonald P., Esteller M., Gray N.S., Haber D.A., Stratton M.R., Benes C.H., Wessels L.F.A., Saez-Rodriguez J., McDermott U., Garnett M.J.
A landscape of pharmacogenomic interactions in cancer.
Cell 166:740-754(2016)
PubMed=28145883; DOI=10.18632/oncotarget.14850
Nouri M., Caradec J., Lubik A.A., Li N., Hollier B.G., Takhar M., Altimirano-Dimas M., Chen M.-Q., Roshan-Moniri M., Butler M., Lehman M., Bishop J., Truong S., Huang S.-C., Cochrane D., Cox M., Collins C., Gleave M.E., Erho N., Alshalafa M., Davicioni E., Nelson C., Gregory-Evans S., Karnes R.J., Jenkins R.B., Klein E.A., Buttyan R.
Therapy-induced developmental reprogramming of prostate cancer cells and acquired therapy resistance.
Oncotarget 8:18949-18967(2017)
PubMed=30305041; DOI=10.1186/s12885-018-4848-x
Jividen K., Kedzierska K.Z., Yang C.-S., Szlachta K., Ratan A., Paschal B.M.
Genomic analysis of DNA repair genes and androgen signaling in prostate cancer.
BMC Cancer 18:960.1-960.20(2018)
PubMed=30244336; DOI=10.1007/s00345-018-2501-6
Samli H., Samli M., Vatansever B., Ardicli S., Aztopal N., Dincel D., Sahin A., Balci F.
Paclitaxel resistance and the role of miRNAs in prostate cancer cell lines.
World J. Urol. 37:1117-1126(2019)
PubMed=30894373; DOI=10.1158/0008-5472.CAN-18-2747
Dutil J., Chen Z.-H., Monteiro A.N.A., Teer J.K., Eschrich S.A.
An interactive resource to probe genetic diversity and estimated ancestry in cancer cell lines.
Cancer Res. 79:1263-1273(2019)
PubMed=31068700; DOI=10.1038/s41586-019-1186-3
Ghandi M., Huang F.W., Jane-Valbuena J., Kryukov G.V., Lo C.C., McDonald E.R. III, Barretina J.G., Gelfand E.T., Bielski C.M., Li H.-X., Hu K., Andreev-Drakhlin A.Y., Kim J., Hess J.M., Haas B.J., Aguet F., Weir B.A., Rothberg M.V., Paolella B.R., Lawrence M.S., Akbani R., Lu Y.-L., Tiv H.L., Gokhale P.C., de Weck A., Mansour A.A., Oh C., Shih J., Hadi K., Rosen Y., Bistline J., Venkatesan K., Reddy A., Sonkin D., Liu M., Lehar J., Korn J.M., Porter D.A., Jones M.D., Golji J., Caponigro G., Taylor J.E., Dunning C.M., Creech A.L., Warren A.C., McFarland J.M., Zamanighomi M., Kauffmann A., Stransky N., Imielinski M., Maruvka Y.E., Cherniack A.D., Tsherniak A., Vazquez F., Jaffe J.D., Lane A.A., Weinstock D.M., Johannessen C.M., Morrissey M.P., Stegmeier F., Schlegel R., Hahn W.C., Getz G., Mills G.B., Boehm J.S., Golub T.R., Garraway L.A., Sellers W.R.
Next-generation characterization of the Cancer Cell Line Encyclopedia.
Nature 569:503-508(2019)
PubMed=31395879; DOI=10.1038/s41467-019-11415-2
Yu K., Chen B., Aran D., Charalel J., Yau C., Wolf D.M., van 't Veer L.J., Butte A.J., Goldstein T., Sirota M.
Comprehensive transcriptomic analysis of cell lines as models of primary tumors across 22 tumor types.
Nat. Commun. 10:3574.1-3574.11(2019)
PubMed=31978347; DOI=10.1016/j.cell.2019.12.023
Nusinow D.P., Szpyt J., Ghandi M., Rose C.M., McDonald E.R. III, Kalocsay M., Jane-Valbuena J., Gelfand E.T., Schweppe D.K., Jedrychowski M.P., Golji J., Porter D.A., Rejtar T., Wang Y.K., Kryukov G.V., Stegmeier F., Erickson B.K., Garraway L.A., Sellers W.R., Gygi S.P.
Quantitative proteomics of the Cancer Cell Line Encyclopedia.
Cell 180:387-402.e16(2020)
PubMed=34402095; DOI=10.1002/pros.24210
Haffner M.C., Bhamidipati A., Tsai H.K., Esopi D.M., Vaghasia A.M., Low J.-Y., Patel R.A., Guner G., Pham M.-T., Castagna N., Hicks J.L., Wyhs N., Aebersold R., De Marzo A.M., Nelson W.G., Guo T.-N., Yegnasubramanian S.
Phenotypic characterization of two novel cell line models of castration-resistant prostate cancer.
Prostate 81:1159-1171(2021)
PubMed=35839778; DOI=10.1016/j.ccell.2022.06.010
Goncalves E., Poulos R.C., Cai Z.-X., Barthorpe S., Manda S.S., Lucas N., Beck A., Bucio-Noble D., Dausmann M., Hall C., Hecker M., Koh J., Lightfoot H., Mahboob S., Mali I., Morris J., Richardson L., Seneviratne A.J., Shepherd R., Sykes E., Thomas F., Valentini S., Williams S.G., Wu Y.-X., Xavier D., MacKenzie K.L., Hains P.G., Tully B., Robinson P.J., Zhong Q., Garnett M.J., Reddel R.R.
Pan-cancer proteomic map of 949 human cell lines.
Cancer Cell 40:835-849.e8(2022)

| 产品名称 | 价格 | 指令 |
| 22RV1人前列腺癌细胞(STR鉴定)[细胞+500ml专培套餐促销] | ¥980 | 购物车 》 |
| DU 145人前列腺癌细胞(STR鉴定)[细胞+500ml专培套餐促销] | ¥980.00 | 购物车 》 |
| LNcapcloneFGC人前列腺癌细胞(STR鉴定)[细胞+500ml专培套餐促销] | ¥980 | 购物车 》 |
| PC-3人前列腺癌细胞(STR鉴定)[细胞+500ml专培套餐促销] | ¥980.00 | 购物车 》 |
| PC-3M人前列腺癌细胞(STR鉴定)[细胞+500ml专培套餐促销] | ¥980.00 | 购物车 》 |
| RM-1小鼠前列腺癌细胞(种属鉴定)[细胞+500ml专培套餐促销] | ¥980.00 | 购物车 》 |
| 国产转染试剂 | ¥800.00 | 购物车 》 |
| 国产优级胎牛血清 | ¥1800.00 | 购物车 》 |
| 荧光素酶标记 | ¥询价 | 购物车 》 |
| 绿/红色荧光蛋白标记 | ¥询价 | 购物车 》 |
| DMEM高糖基础培养基 | ¥56.00 | 购物车 》 |
| DMEM高糖完全培养基(10%FBS) | ¥350.00 | 购物车 》 |
| 人源细胞STR鉴定服务 | ¥900.00 | 购物车 》 |
 上海中乔新舟生物科技有限公司
上海中乔新舟生物科技有限公司