|
产品名称 |
Jurkat,Clone E6-1人T淋巴细胞白血病细胞 |
|
货号 |
ZQ0083 |
|
产品介绍 |
Jurkat,Clone E6-1是一种人T淋巴细胞白血病细胞系,Jurkat细胞由Schneider建立,来源于一个14岁的男孩的外周血。该克隆是 Jurkat-FHCRC细胞株的一个克隆(Jurkat的一个衍生株),这个细胞系具有T淋巴细胞的特性,并且在佛波酯(phorbol esters)和外源凝集素(lectins)或抗T3单克隆抗体的刺激下能够产生大量的白细胞介素-2(IL-2),使其成为研究T细胞功能和信号传导的重要模型。在免疫学研究领域得到了广泛的应用,尤其是在T细胞受体信号传导、细胞因子产生、免疫细胞间的相互作用以及免疫疾病的分子机制研究中具有重要价值。 备注: 细胞对血清质量较为敏感,我司建议您使用进口大品牌优质血清进行培养。该细胞对细胞密度较为敏感,培养、传代时请注意保持细胞密度在合适的范围。
我司提供Jurkat, Clone E6-1细胞无血清专用培养基,欢迎咨询订购。 |
|
种属 |
人 |
|
性别/年龄 |
男/14岁 |
|
组织 |
外周血 |
|
疾病 |
急性T细胞白血病 |
|
细胞类型 |
肿瘤细胞 |
|
形态学 |
淋巴母细胞 |
|
生长方式 |
悬浮 |
|
倍增时间 |
大约48~72小时 |
|
常规培养基和添加剂 |
RPMI-1640(中乔新舟 货号:ZQ-200)+10%胎牛血清(中乔新舟 货号:AU0600)+1%双抗(中乔新舟 货号:CSP006) |
|
推荐含血清完全培养基货号 |
|
|
推荐无血清完全培养基货号 |
|
|
生物安全等级 |
BSL-1 |
|
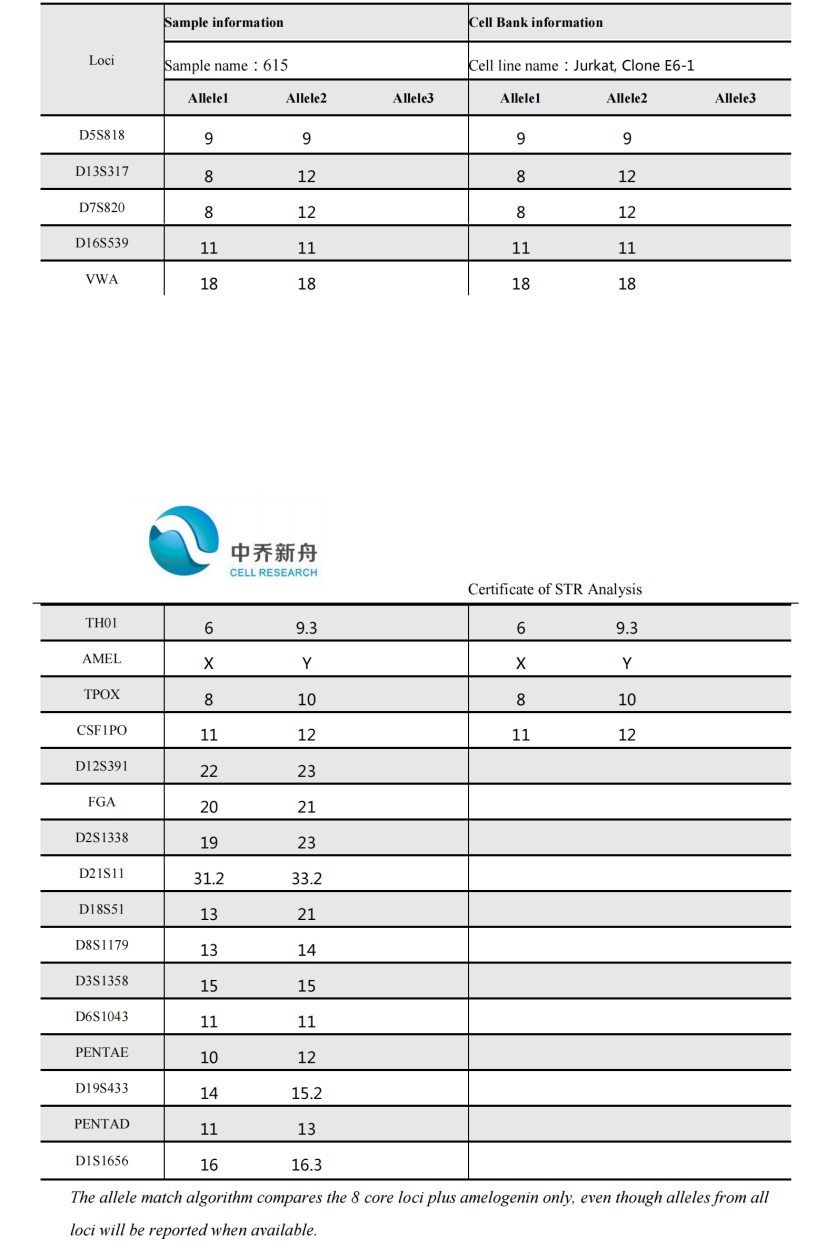
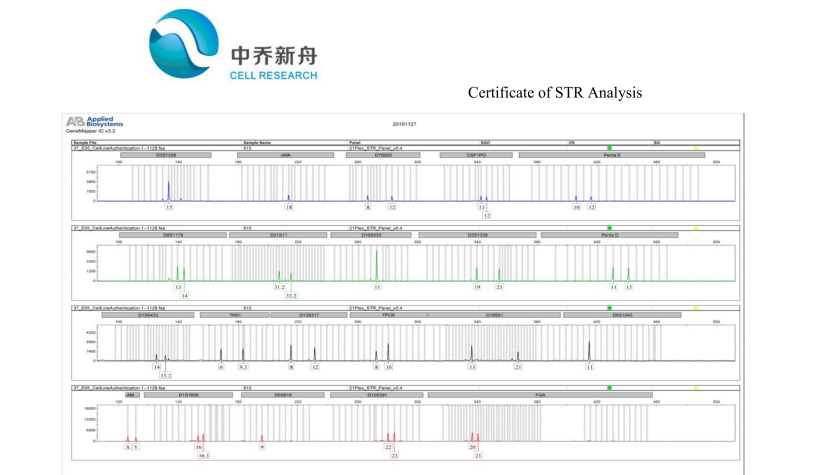
STR位点信息 |
Amelogenin: X,Y
|
|
培养条件 |
95%空气,5%二氧化碳;37℃ |
|
抗原表达/受体表达 |
*** |
|
基因表达 |
*** |
|
保藏机构 |
ATCC; TIB-152 BCRC; 60424 BCRJ; 0125 ECACC; 88042803 |
|
供应限制 |
仅供科研使用 |
|
货号 |
ZQ0083 |
|
发货规格 |
活细胞:T25培养瓶*1瓶或者1ml 冻存管*2支(细胞量约为1x10^6 Cells/Vial)二选一 |
|
发货形式 |
活细胞:常温运输;冻存管:干冰运输 |
|
储存温度 |
活细胞:培养箱;冻存管:液氮罐 |
|
产地 |
中国 |
|
供应限制 |
仅供科研使用 |
| 货号 | 产品名称 | 规格 | 价格 | 指令 |
 上海中乔新舟生物科技有限公司
上海中乔新舟生物科技有限公司