|
产品名称 |
HCT-15人结直肠腺癌细胞 |
|
货号 |
ZQ0324 |
|
产品介绍 |
HCT-15细胞来源于一名男性的结肠腺癌。该细胞系于20世纪70年代初被发现,广泛应用于癌症研究领域,特别是探索结直肠癌的生物学和治疗。DNA fingerprinting证据表明,这株细胞和DLD-1 (ATCC CCL-221) 来源于同一个人;但同工酶及细胞染色体组型分析仍存疑问。 细胞呈CSAp阴性(CSAp-)。 角蛋白免疫过氧化物酶染色阳性。
注意事项: |
|
种属 |
人 |
|
性别/年龄 |
男 |
|
组织 |
结肠 |
|
疾病 |
杜克斯氏C型,结肠直肠腺癌 |
|
细胞类型 |
肿瘤细胞 |
|
形态学 |
上皮 |
|
生长方式 |
贴壁 |
|
倍增时间 |
大约19~29.57小时 |
|
培养基和添加剂 |
RPMI-1640(中乔新舟 货号:ZQ-200)+10%胎牛血清(中乔新舟 货号:ZQ500-A)+1%P/S(中乔新舟 货号:CSP006) |
|
推荐完全培养基货号 |
|
|
生物安全等级 |
BSL-1 |
|
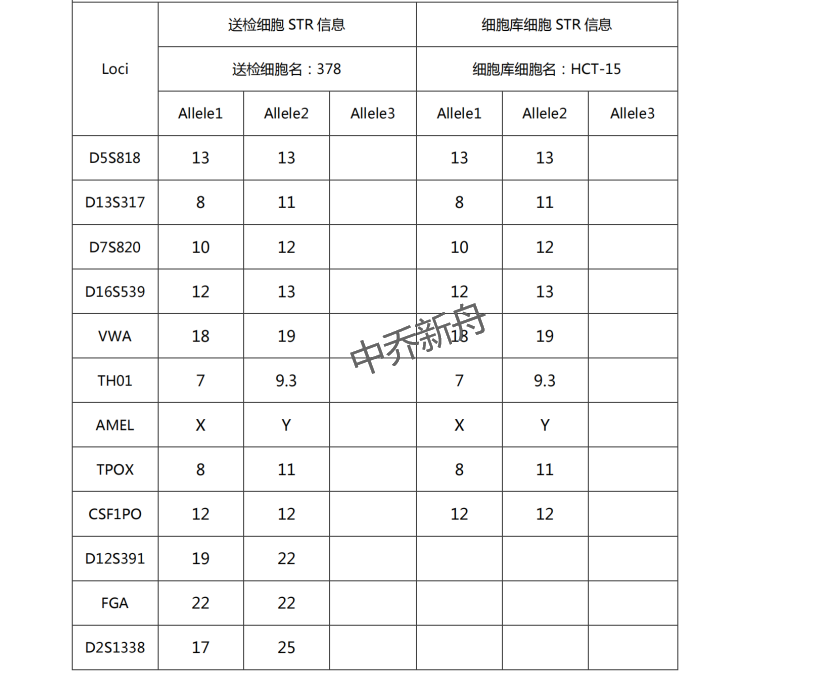
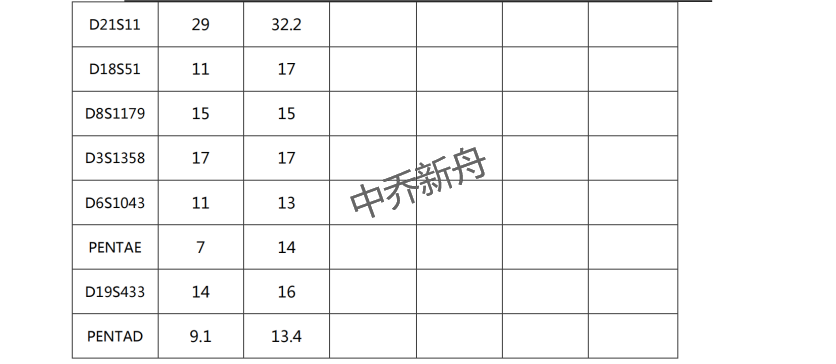
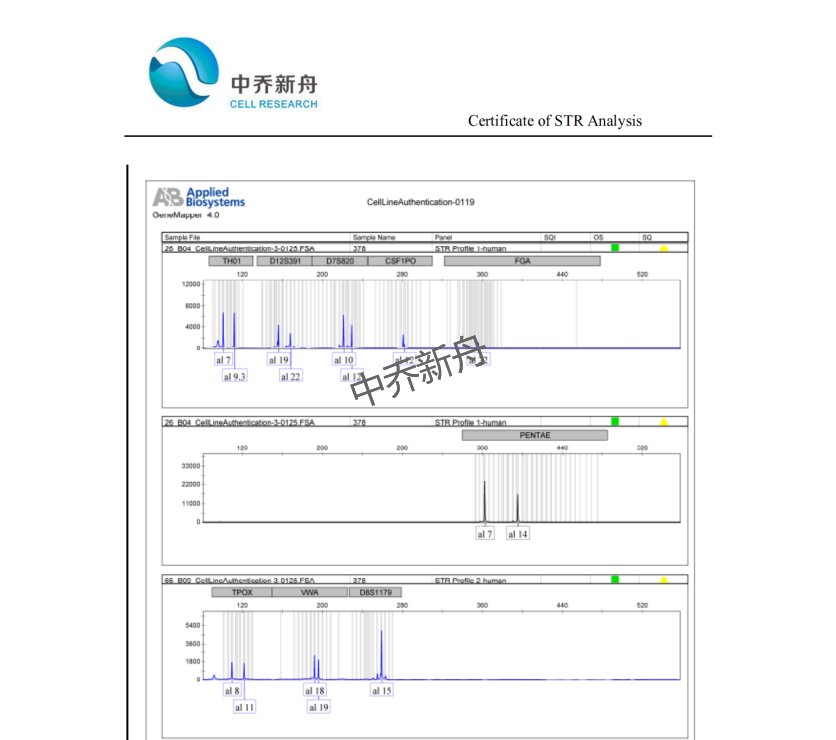
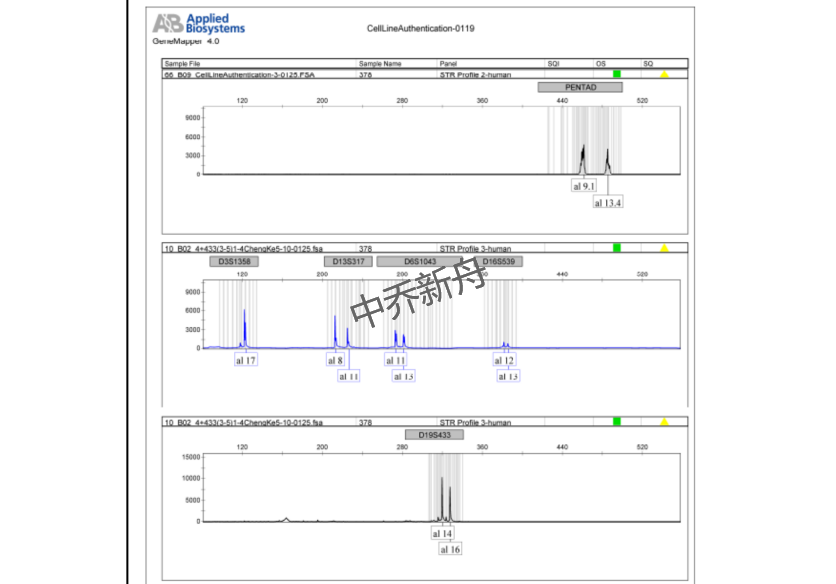
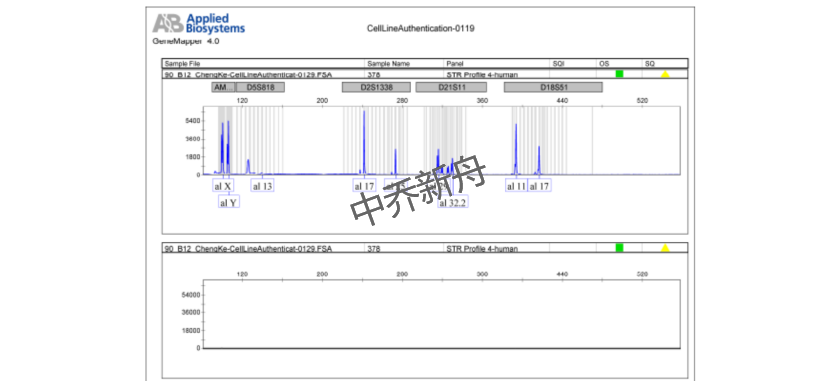
STR位点信息 |
Amelogenin: X,Y |
|
培养条件 |
95%空气,5%二氧化碳;37℃ |
|
抗原表达/受体表达 |
*** |
|
基因表达 |
*** |
|
保藏机构 |
ATCC; CCL-225 BCRC; 60539 DSMZ; ACC-357 ECACC; 91030712 |
|
供应限制 |
仅供科研使用 |
|
货号 |
ZQ0324 |
|
发货规格 |
活细胞:T25培养瓶*1瓶或者1ml 冻存管*2支(细胞量约为1x10^6 Cells/Vial)二选一 |
|
发货形式 |
活细胞:常温运输;冻存管:干冰运输 |
|
储存温度 |
活细胞:培养箱;冻存管:液氮罐 |
|
产地 |
中国 |
|
供应限制 |
仅供科研使用 |
PubMed=427742
Dexter D.L., Barbosa J.A., Calabresi P.
N,N-dimethylformamide-induced alteration of cell culture characteristics and loss of tumorigenicity in cultured human colon carcinoma cells.
Cancer Res. 39:1020-1025(1979)
PubMed=7247963; DOI=10.1016/0006-2952(81)90167-2
Crabtree G.W., Dexter D.L., Stoeckler J.D., Savarese T.M., Ghoda L.Y., Rogler-Brown T.L., Calabresi P., Parks R.E. Jr.
Activities of purine-metabolizing enzymes in human colon carcinoma cell lines and xenograft tumors.
Biochem. Pharmacol. 30:793-798(1981)
PubMed=7139607
Dexter D.L., Spremulli E.N., Matook G.M., Diamond I., Calabresi P.
Inhibition of the growth of human colon cancer xenografts by polar solvents.
Cancer Res. 42:5018-5022(1982)
PubMed=2041050; DOI=10.1093/jnci/83.11.757
Monks A., Scudiero D.A., Skehan P., Shoemaker R.H., Paull K.D., Vistica D.T., Hose C.D., Langley J., Cronise P., Vaigro-Wolff A., Gray-Goodrich M., Campbell H., Mayo J.G., Boyd M.R.
Feasibility of a high-flux anticancer drug screen using a diverse panel of cultured human tumor cell lines.
J. Natl. Cancer Inst. 83:757-766(1991)
PubMed=8464898; DOI=10.1073/pnas.90.7.2842
Browning M.J., Krausa P., Rowan A.J., Bicknell D.C., Bodmer J.G., Bodmer W.F.
Tissue typing the HLA-A locus from genomic DNA by sequence-specific PCR: comparison of HLA genotype and surface expression on colorectal tumor cell lines.
Proc. Natl. Acad. Sci. U.S.A. 90:2842-2845(1993)
PubMed=7972006; DOI=10.1073/pnas.91.23.11045
Okamoto A., Demetrick D.J., Spillare E.A., Hagiwara K., Hussain S.P., Bennett W.P., Forrester K., Gerwin B.I., Serrano M., Beach D.H., Harris C.C.
Mutations and altered expression of p16INK4 in human cancer.
Proc. Natl. Acad. Sci. U.S.A. 91:11045-11049(1994)
PubMed=8197130; DOI=10.1073/pnas.91.11.4751
Bicknell D.C., Rowan A.J., Bodmer W.F.
Beta 2-microglobulin gene mutations: a study of established colorectal cell lines and fresh tumors.
Proc. Natl. Acad. Sci. U.S.A. 91:4751-4755(1994)
PubMed=7621404; DOI=10.1016/0165-4608(94)00225-z
Chen T.R., Dorotinsky C.S., McGuire L.J., Macy M.L., Hay R.J.
DLD-1 and HCT-15 cell lines derived separately from colorectal carcinomas have totally different chromosome changes but the same genetic origin.
Cancer Genet. Cytogenet. 81:103-108(1995)
PubMed=9000147
Cottu P.-H., Muzeau F., Estreicher A., Flejou J.-F., Iggo R.D., Thomas G., Hamelin R.
Inverse correlation between RER+ status and p53 mutation in colorectal cancer cell lines.
Oncogene 13:2727-2730(1996)
PubMed=9000572
Hoang J.-M., Cottu P.-H., Thuille B., Salmon R.J., Thomas G., Hamelin R.
BAT-26, an indicator of the replication error phenotype in colorectal cancers and cell lines.
Cancer Res. 57:300-303(1997)
PubMed=9809040; DOI=10.1016/S0165-4608(98)00081-8
Vermeulen S.J., Chen T.-R., Speleman F., Nollet F., Van Roy F.M., Mareel M.M.
Did the four human cancer cell lines DLD-1, HCT-15, HCT-8, and HRT-18 originate from one and the same patient?
Cancer Genet. Cytogenet. 107:76-79(1998)
PubMed=10700174; DOI=10.1038/73432
Ross D.T., Scherf U., Eisen M.B., Perou C.M., Rees C., Spellman P.T., Iyer V.R., Jeffrey S.S., van de Rijn M., Waltham M.C., Pergamenschikov A., Lee J.C.F., Lashkari D., Shalon D., Myers T.G., Weinstein J.N., Botstein D., Brown P.O.
Systematic variation in gene expression patterns in human cancer cell lines.
Nat. Genet. 24:227-235(2000)
PubMed=10737795; DOI=10.1073/pnas.97.7.3352
Rowan A.J., Lamlum H., Ilyas M., Wheeler J., Straub J., Papadopoulou A., Bicknell D.C., Bodmer W.F., Tomlinson I.P.M.
APC mutations in sporadic colorectal tumors: a mutational 'hotspot' and interdependence of the 'two hits'.
Proc. Natl. Acad. Sci. U.S.A. 97:3352-3357(2000)
PubMed=11314036; DOI=10.1038/sj.onc.1204211
Forgacs E., Wren J.D., Kamibayashi C., Kondo M., Xu X.L., Markowitz S.D., Tomlinson G.E., Muller C.Y., Gazdar A.F., Garner H.R., Minna J.D.
Searching for microsatellite mutations in coding regions in lung, breast, ovarian and colorectal cancers.
Oncogene 20:1005-1009(2001)
PubMed=11414198; DOI=10.1007/s004320000207
Lahm H., Andre S., Hoeflich A., Fischer J.R., Sordat B., Kaltner H., Wolf E., Gabius H.-J.
Comprehensive galectin fingerprinting in a panel of 61 human tumor cell lines by RT-PCR and its implications for diagnostic and therapeutic procedures.
J. Cancer Res. Clin. Oncol. 127:375-386(2001)
PubMed=11416159; DOI=10.1073/pnas.121616198
Masters J.R.W., Thomson J.A., Daly-Burns B., Reid Y.A., Dirks W.G., Packer P., Toji L.H., Ohno T., Tanabe H., Arlett C.F., Kelland L.R., Harrison M., Virmani A.K., Ward T.H., Ayres K.L., Debenham P.G.
Short tandem repeat profiling provides an international reference standard for human cell lines.
Proc. Natl. Acad. Sci. U.S.A. 98:8012-8017(2001)
PubMed=11526487; DOI=10.1038/sj.onc.1204611
Gayet J., Zhou X.-P., Duval A., Rolland S., Hoang J.-M., Cottu P.-H., Hamelin R.
Extensive characterization of genetic alterations in a series of human colorectal cancer cell lines.
Oncogene 20:5025-5032(2001)
PubMed=11668190; DOI=10.1177/002215540104901105
Quentmeier H., Osborn M., Reinhardt J., Zaborski M., Drexler H.G.
Immunocytochemical analysis of cell lines derived from solid tumors.
J. Histochem. Cytochem. 49:1369-1378(2001)
PubMed=12584437; DOI=10.1159/000068544
Melcher R., Koehler S., Steinlein C., Schmid M., Mueller C.R., Luehrs H., Menzel T., Scheppach W., Moerk H., Scheurlen M., Koehrle J., Al-Taie O.
Spectral karyotype analysis of colon cancer cell lines of the tumor suppressor and mutator pathway.
Cytogenet. Genome Res. 98:22-28(2002)
PubMed=15748285; DOI=10.1186/1479-5876-3-11
Adams S., Robbins F.-M., Chen D., Wagage D., Holbeck S.L., Morse H.C. III, Stroncek D., Marincola F.M.
HLA class I and II genotype of the NCI-60 cell lines.
J. Transl. Med. 3:11.1-11.8(2005)
PubMed=15900046; DOI=10.1093/jnci/dji133
Mashima T., Oh-hara T., Sato S., Mochizuki M., Sugimoto Y., Yamazaki K., Hamada J.-i., Tada M., Moriuchi T., Ishikawa Y., Kato Y., Tomoda H., Yamori T., Tsuruo T.
p53-defective tumors with a functional apoptosome-mediated pathway: a new therapeutic target.
J. Natl. Cancer Inst. 97:765-777(2005)
PubMed=16854228; DOI=10.1186/1476-4598-5-29
Bandres Elizalde E.M., Cubedo E., Agirre X., Malumbres R., Zarate R., Ramirez N., Abajo A., Navarro A., Moreno I., Monzo M., Garcia-Foncillas J.
Identification by real-time PCR of 13 mature microRNAs differentially expressed in colorectal cancer and non-tumoral tissues.
Mol. Cancer 5:29.1-29.10(2006)
PubMed=17088437; DOI=10.1158/1535-7163.MCT-06-0433
Ikediobi O.N., Davies H., Bignell G.R., Edkins S., Stevens C., O'Meara S., Santarius T., Avis T., Barthorpe S., Brackenbury L., Buck G., Butler A.P., Clements J., Cole J., Dicks E., Forbes S., Gray K., Halliday K., Harrison R., Hills K., Hinton J., Hunter C., Jenkinson A., Jones D., Kosmidou V., Lugg R., Menzies A., Mironenko T., Parker A., Perry J., Raine K.M., Richardson D., Shepherd R., Small A., Smith R., Solomon H., Stephens P.J., Teague J.W., Tofts C., Varian J., Webb T., West S., Widaa S., Yates A., Reinhold W.C., Weinstein J.N., Stratton M.R., Futreal P.A., Wooster R.
Mutation analysis of 24 known cancer genes in the NCI-60 cell line set.
Mol. Cancer Ther. 5:2606-2612(2006)
PubMed=18167186; DOI=10.5555/cmj.0366-6999.120.23.p2119.01
Liu W.-D., Zhong B.-Y., Zhang Y.-D., Choi G.-S.
Mutation analysis of the checkpoint kinase 2 gene in colorectal cancer cell lines.
Chin. Med. J. 120:2119-2123(2007)
PubMed=19372543; DOI=10.1158/1535-7163.MCT-08-0921
Lorenzi P.L., Reinhold W.C., Varma S., Hutchinson A.A., Pommier Y., Chanock S.J., Weinstein J.N.
DNA fingerprinting of the NCI-60 cell line panel.
Mol. Cancer Ther. 8:713-724(2009)
PubMed=20164919; DOI=10.1038/nature08768
Bignell G.R., Greenman C.D., Davies H., Butler A.P., Edkins S., Andrews J.M., Buck G., Chen L., Beare D., Latimer C., Widaa S., Hinton J., Fahey C., Fu B.-Y., Swamy S., Dalgliesh G.L., Teh B.T., Deloukas P., Yang F.-T., Campbell P.J., Futreal P.A., Stratton M.R.
Signatures of mutation and selection in the cancer genome.
Nature 463:893-898(2010)
PubMed=20215515; DOI=10.1158/0008-5472.CAN-09-3458
Rothenberg S.M., Mohapatra G., Rivera M.N., Winokur D., Greninger P., Nitta M., Sadow P.M., Sooriyakumar G., Brannigan B.W., Ulman M.J., Perera R.M., Wang R., Tam A., Ma X.-J., Erlander M., Sgroi D.C., Rocco J.W., Lingen M.W., Cohen E.E.W., Louis D.N., Settleman J., Haber D.A.
A genome-wide screen for microdeletions reveals disruption of polarity complex genes in diverse human cancers.
Cancer Res. 70:2158-2164(2010)
PubMed=20570890; DOI=10.1158/0008-5472.CAN-10-0192
Janakiraman M., Vakiani E., Zeng Z.-S., Pratilas C.A., Taylor B.S., Chitale D., Halilovic E., Wilson M., Huberman K., Ricarte Filho J.C.M., Persaud Y., Levine D.A., Fagin J.A., Jhanwar S.C., Mariadason J.M., Lash A., Ladanyi M., Saltz L.B., Heguy A., Paty P.B., Solit D.B.
Genomic and biological characterization of exon 4 KRAS mutations in human cancer.
Cancer Res. 70:5901-5911(2010)
PubMed=20606684; DOI=10.1038/sj.bjc.6605780
Bracht K., Nicholls A.M., Liu Y., Bodmer W.F.
5-fluorouracil response in a large panel of colorectal cancer cell lines is associated with mismatch repair deficiency.
Br. J. Cancer 103:340-346(2010)
PubMed=22068913; DOI=10.1073/pnas.1111840108
Gillet J.-P., Calcagno A.M., Varma S., Marino M., Green L.J., Vora M.I., Patel C., Orina J.N., Eliseeva T.A., Singal V., Padmanabhan R., Davidson B., Ganapathi R., Sood A.K., Rueda B.R., Ambudkar S.V., Gottesman M.M.
Redefining the relevance of established cancer cell lines to the study of mechanisms of clinical anti-cancer drug resistance.
Proc. Natl. Acad. Sci. U.S.A. 108:18708-18713(2011)
PubMed=22336246; DOI=10.1016/j.bmc.2012.01.017
Kong D.-X., Yamori T.
JFCR39, a panel of 39 human cancer cell lines, and its application in the discovery and development of anticancer drugs.
Bioorg. Med. Chem. 20:1947-1951(2012)
PubMed=22347499; DOI=10.1371/journal.pone.0031628
Ruan X.-Y., Kocher J.-P.A., Pommier Y., Liu H.-F., Reinhold W.C.
Mass homozygotes accumulation in the NCI-60 cancer cell lines as compared to HapMap trios, and relation to fragile site location.
PLoS ONE 7:E31628-E31628(2012)
PubMed=22384151; DOI=10.1371/journal.pone.0032096
Lee J.-S., Kim Y.K., Kim H.J., Hajar S., Tan Y.L., Kang N.-Y., Ng S.H., Yoon C.N., Chang Y.-T.
Identification of cancer cell-line origins using fluorescence image-based phenomic screening.
PLoS ONE 7:E32096-E32096(2012)
PubMed=22460905; DOI=10.1038/nature11003
Barretina J.G., Caponigro G., Stransky N., Venkatesan K., Margolin A.A., Kim S., Wilson C.J., Lehar J., Kryukov G.V., Sonkin D., Reddy A., Liu M., Murray L., Berger M.F., Monahan J.E., Morais P., Meltzer J., Korejwa A., Jane-Valbuena J., Mapa F.A., Thibault J., Bric-Furlong E., Raman P., Shipway A., Engels I.H., Cheng J., Yu G.-Y.K., Yu J.-J., Aspesi P. Jr., de Silva M., Jagtap K., Jones M.D., Wang L., Hatton C., Palescandolo E., Gupta S., Mahan S., Sougnez C., Onofrio R.C., Liefeld T., MacConaill L.E., Winckler W., Reich M., Li N.-X., Mesirov J.P., Gabriel S.B., Getz G., Ardlie K., Chan V., Myer V.E., Weber B.L., Porter J., Warmuth M., Finan P., Harris J.L., Meyerson M.L., Golub T.R., Morrissey M.P., Sellers W.R., Schlegel R., Garraway L.A.
The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity.
Nature 483:603-607(2012)
PubMed=22628656; DOI=10.1126/science.1218595
Jain M., Nilsson R., Sharma S., Madhusudhan N., Kitami T., Souza A.L., Kafri R., Kirschner M.W., Clish C.B., Mootha V.K.
Metabolite profiling identifies a key role for glycine in rapid cancer cell proliferation.
Science 336:1040-1044(2012)
PubMed=23856246; DOI=10.1158/0008-5472.CAN-12-3342
Abaan O.D., Polley E.C., Davis S.R., Zhu Y.-L.J., Bilke S., Walker R.L., Pineda M.A., Gindin Y., Jiang Y., Reinhold W.C., Holbeck S.L., Simon R.M., Doroshow J.H., Pommier Y., Meltzer P.S.
The exomes of the NCI-60 panel: a genomic resource for cancer biology and systems pharmacology.
Cancer Res. 73:4372-4382(2013)
PubMed=23933261; DOI=10.1016/j.celrep.2013.07.018
Moghaddas Gholami A., Hahne H., Wu Z.-X., Auer F.J., Meng C., Wilhelm M., Kuster B.
Global proteome analysis of the NCI-60 cell line panel.
Cell Rep. 4:609-620(2013)
PubMed=24042735; DOI=10.1038/oncsis.2013.35
Ahmed D., Eide P.W., Eilertsen I.A., Danielsen S.A., Eknaes M., Hektoen M., Lind G.E., Lothe R.A.
Epigenetic and genetic features of 24 colon cancer cell lines.
Oncogenesis 2:e71.1-e71.8(2013)
PubMed=24279929; DOI=10.1186/2049-3002-1-20
Dolfi S.C., Chan L.L.-Y., Qiu J., Tedeschi P.M., Bertino J.R., Hirshfield K.M., Oltvai Z.N., Vazquez A.
The metabolic demands of cancer cells are coupled to their size and protein synthesis rates.
Cancer Metab. 1:20.1-20.13(2013)
PubMed=24670534; DOI=10.1371/journal.pone.0092047
Varma S., Pommier Y., Sunshine M., Weinstein J.N., Reinhold W.C.
High resolution copy number variation data in the NCI-60 cancer cell lines from whole genome microarrays accessible through CellMiner.
PLoS ONE 9:E92047-E92047(2014)
PubMed=24755471; DOI=10.1158/0008-5472.CAN-14-0013
Mouradov D., Sloggett C., Jorissen R.N., Love C.G., Li S., Burgess A.W., Arango D., Strausberg R.L., Buchanan D., Wormald S., O'Connor L., Wilding J.L., Bicknell D.C., Tomlinson I.P.M., Bodmer W.F., Mariadason J.M., Sieber O.M.
Colorectal cancer cell lines are representative models of the main molecular subtypes of primary cancer.
Cancer Res. 74:3238-3247(2014)
PubMed=25485619; DOI=10.1038/nbt.3080
Klijn C., Durinck S., Stawiski E.W., Haverty P.M., Jiang Z.-S., Liu H.-B., Degenhardt J., Mayba O., Gnad F., Liu J.-F., Pau G., Reeder J., Cao Y., Mukhyala K., Selvaraj S.K., Yu M.-M., Zynda G.J., Brauer M.J., Wu T.D., Gentleman R.C., Manning G., Yauch R.L., Bourgon R., Stokoe D., Modrusan Z., Neve R.M., de Sauvage F.J., Settleman J., Seshagiri S., Zhang Z.-M.
A comprehensive transcriptional portrait of human cancer cell lines.
Nat. Biotechnol. 33:306-312(2015)
PubMed=25877200; DOI=10.1038/nature14397
Yu M., Selvaraj S.K., Liang-Chu M.M.Y., Aghajani S., Busse M., Yuan J., Lee G., Peale F.V., Klijn C., Bourgon R., Kaminker J.S., Neve R.M.
A resource for cell line authentication, annotation and quality control.
Nature 520:307-311(2015)
PubMed=25926053; DOI=10.1038/ncomms8002
Medico E., Russo M., Picco G., Cancelliere C., Valtorta E., Corti G., Buscarino M., Isella C., Lamba S., Martinoglio B., Veronese S., Siena S., Sartore-Bianchi A., Beccuti M., Mottolese M., Linnebacher M., Cordero F., Di Nicolantonio F., Bardelli A.
The molecular landscape of colorectal cancer cell lines unveils clinically actionable kinase targets.
Nat. Commun. 6:7002.1-7002.10(2015)
PubMed=25944804; DOI=10.1158/1078-0432.CCR-14-2457
Bazzocco S., Dopeso H., Carton-Garcia F., Macaya I., Andretta E., Chionh F., Rodrigues P., Garrido M., Alazzouzi H., Nieto R., Sanchez A., Schwartz S. Jr., Bilic J., Mariadason J.M., Arango D.
Highly expressed genes in rapidly proliferating tumor cells as new targets for colorectal cancer treatment.
Clin. Cancer Res. 21:3695-3704(2015)
PubMed=26169745; DOI=10.1186/s12967-015-0576-z
Halama A., Guerrouahen B.S., Pasquier J., Diboun I., Karoly E.D., Suhre K., Rafii A.
Metabolic signatures differentiate ovarian from colon cancer cell lines.
J. Transl. Med. 13:223.1-223.12(2015)
PubMed=26589293; DOI=10.1186/s13073-015-0240-5
Scholtalbers J., Boegel S., Bukur T., Byl M., Goerges S., Sorn P., Loewer M., Sahin U., Castle J.C.
TCLP: an online cancer cell line catalogue integrating HLA type, predicted neo-epitopes, virus and gene expression.
Genome Med. 7:118.1-118.7(2015)
PubMed=26537799; DOI=10.1074/mcp.M115.051235
Holst S., Deuss A.J.M., van Pelt G.W., van Vliet S.J., Garcia-Vallejo J.J., Koeleman C.A.M., Deelder A.M., Mesker W.E., Tollenaar R.A.E.M., Rombouts Y., Wuhrer M.
N-glycosylation profiling of colorectal cancer cell lines reveals association of fucosylation with differentiation and caudal type homebox 1 (CDX1)/villin mRNA expression.
Mol. Cell. Proteomics 15:124-140(2016)
PubMed=27377824; DOI=10.1038/sdata.2016.52
Mestdagh P., Lefever S., Volders P.-J., Derveaux S., Hellemans J., Vandesompele J.
Long non-coding RNA expression profiling in the NCI60 cancer cell line panel using high-throughput RT-qPCR.
Sci. Data 3:160052-160052(2016)
PubMed=27397505; DOI=10.1016/j.cell.2016.06.017
Iorio F., Knijnenburg T.A., Vis D.J., Bignell G.R., Menden M.P., Schubert M., Aben N., Goncalves E., Barthorpe S., Lightfoot H., Cokelaer T., Greninger P., van Dyk E., Chang H., de Silva H., Heyn H., Deng X.-M., Egan R.K., Liu Q.-S., Mironenko T., Mitropoulos X., Richardson L., Wang J.-H., Zhang T.-H., Moran S., Sayols S., Soleimani M., Tamborero D., Lopez-Bigas N., Ross-Macdonald P., Esteller M., Gray N.S., Haber D.A., Stratton M.R., Benes C.H., Wessels L.F.A., Saez-Rodriguez J., McDermott U., Garnett M.J.
A landscape of pharmacogenomic interactions in cancer.
Cell 166:740-754(2016)
PubMed=27807467; DOI=10.1186/s13100-016-0078-4
Zampella J.G., Rodic N., Yang W.R., Huang C.R.L., Welch J., Gnanakkan V.P., Cornish T.C., Boeke J.D., Burns K.H.
A map of mobile DNA insertions in the NCI-60 human cancer cell panel.
Mob. DNA 7:20.1-20.11(2016)
PubMed=28179481; DOI=10.1158/1535-7163.MCT-16-0578
Tanaka N., Mashima T., Mizutani A., Sato A., Aoyama A., Gong B., Yoshida H., Muramatsu Y., Nakata K., Matsuura M., Katayama R., Nagayama S., Fujita N., Sugimoto Y., Seimiya H.
APC mutations as a potential biomarker for sensitivity to tankyrase inhibitors in colorectal cancer.
Mol. Cancer Ther. 16:752-762(2017)
PubMed=28192450; DOI=10.1371/journal.pone.0171435
Fasterius E., Raso C., Kennedy S.A., Rauch N., Lundin P., Kolch W., Uhlen M., Al-Khalili Szigyarto C.
A novel RNA sequencing data analysis method for cell line authentication.
PLoS ONE 12:E0171435-E0171435(2017)
PubMed=28196595; DOI=10.1016/j.ccell.2017.01.005
Li J., Zhao W., Akbani R., Liu W.-B., Ju Z.-L., Ling S.-Y., Vellano C.P., Roebuck P., Yu Q.-H., Eterovic A.K., Byers L.A., Davies M.A., Deng W.-L., Gopal Y.N.V., Chen G., von Euw E.M., Slamon D.J., Conklin D., Heymach J.V., Gazdar A.F., Minna J.D., Myers J.N., Lu Y.-L., Mills G.B., Liang H.
Characterization of human cancer cell lines by reverse-phase protein arrays.
Cancer Cell 31:225-239(2017)
PubMed=28683746; DOI=10.1186/s12943-017-0691-y
Berg K.C.G., Eide P.W., Eilertsen I.A., Johannessen B., Bruun J., Danielsen S.A., Bjornslett M., Meza-Zepeda L.A., Eknaes M., Lind G.E., Myklebost O., Skotheim R.I., Sveen A., Lothe R.A.
Multi-omics of 34 colorectal cancer cell lines -- a resource for biomedical studies.
Mol. Cancer 16:116.1-116.16(2017)
PubMed=28854368; DOI=10.1016/j.celrep.2017.08.010
Roumeliotis T.I., Williams S.P., Goncalves E., Alsinet C., Del Castillo Velasco-Herrera M., Aben N., Ghavidel F.Z., Michaut M., Schubert M., Price S., Wright J.C., Yu L., Yang M., Dienstmann R., Guinney J., Beltrao P., Brazma A., Pardo M., Stegle O., Adams D.J., Wessels L.F.A., Saez-Rodriguez J., McDermott U., Choudhary J.S.
Genomic determinants of protein abundance variation in colorectal cancer cells.
Cell Rep. 20:2201-2214(2017)
PubMed=29444439; DOI=10.1016/j.celrep.2018.01.051
Yuan T.L., Amzallag A., Bagni R., Yi M., Afghani S., Burgan W., Fer N., Strathern L.A., Powell K., Smith B., Waters A.M., Drubin D.A., Thomson T., Liao R., Greninger P., Stein G.T., Murchie E., Cortez E., Egan R.K., Procter L., Bess M., Cheng K.T., Lee C.-S., Lee L.C., Fellmann C., Stephens R., Luo J., Lowe S.W., Benes C.H., McCormick F.
Differential effector engagement by oncogenic KRAS.
Cell Rep. 22:1889-1902(2018)
PubMed=30894373; DOI=10.1158/0008-5472.CAN-18-2747
Dutil J., Chen Z.-H., Monteiro A.N.A., Teer J.K., Eschrich S.A.
An interactive resource to probe genetic diversity and estimated ancestry in cancer cell lines.
Cancer Res. 79:1263-1273(2019)
PubMed=30971826; DOI=10.1038/s41586-019-1103-9
Behan F.M., Iorio F., Picco G., Goncalves E., Beaver C.M., Migliardi G., Santos R., Rao Y., Sassi F., Pinnelli M., Ansari R., Harper S., Jackson D.A., McRae R., Pooley R., Wilkinson P., van der Meer D.J., Dow D., Buser-Doepner C.A., Bertotti A., Trusolino L., Stronach E.A., Saez-Rodriguez J., Yusa K., Garnett M.J.
Prioritization of cancer therapeutic targets using CRISPR-Cas9 screens.
Nature 568:511-516(2019)
PubMed=31068700; DOI=10.1038/s41586-019-1186-3
Ghandi M., Huang F.W., Jane-Valbuena J., Kryukov G.V., Lo C.C., McDonald E.R. III, Barretina J.G., Gelfand E.T., Bielski C.M., Li H.-X., Hu K., Andreev-Drakhlin A.Y., Kim J., Hess J.M., Haas B.J., Aguet F., Weir B.A., Rothberg M.V., Paolella B.R., Lawrence M.S., Akbani R., Lu Y.-L., Tiv H.L., Gokhale P.C., de Weck A., Mansour A.A., Oh C., Shih J., Hadi K., Rosen Y., Bistline J., Venkatesan K., Reddy A., Sonkin D., Liu M., Lehar J., Korn J.M., Porter D.A., Jones M.D., Golji J., Caponigro G., Taylor J.E., Dunning C.M., Creech A.L., Warren A.C., McFarland J.M., Zamanighomi M., Kauffmann A., Stransky N., Imielinski M., Maruvka Y.E., Cherniack A.D., Tsherniak A., Vazquez F., Jaffe J.D., Lane A.A., Weinstock D.M., Johannessen C.M., Morrissey M.P., Stegmeier F., Schlegel R., Hahn W.C., Getz G., Mills G.B., Boehm J.S., Golub T.R., Garraway L.A., Sellers W.R.
Next-generation characterization of the Cancer Cell Line Encyclopedia.
Nature 569:503-508(2019)
PubMed=31978347; DOI=10.1016/j.cell.2019.12.023
Nusinow D.P., Szpyt J., Ghandi M., Rose C.M., McDonald E.R. III, Kalocsay M., Jane-Valbuena J., Gelfand E.T., Schweppe D.K., Jedrychowski M.P., Golji J., Porter D.A., Rejtar T., Wang Y.K., Kryukov G.V., Stegmeier F., Erickson B.K., Garraway L.A., Sellers W.R., Gygi S.P.
Quantitative proteomics of the Cancer Cell Line Encyclopedia.
Cell 180:387-402.e16(2020)
PubMed=35839778; DOI=10.1016/j.ccell.2022.06.010
Goncalves E., Poulos R.C., Cai Z.-X., Barthorpe S., Manda S.S., Lucas N., Beck A., Bucio-Noble D., Dausmann M., Hall C., Hecker M., Koh J., Lightfoot H., Mahboob S., Mali I., Morris J., Richardson L., Seneviratne A.J., Shepherd R., Sykes E., Thomas F., Valentini S., Williams S.G., Wu Y.-X., Xavier D., MacKenzie K.L., Hains P.G., Tully B., Robinson P.J., Zhong Q., Garnett M.J., Reddel R.R.
Pan-cancer proteomic map of 949 human cell lines.
Cancer Cell 40:835-849.e8(2022)
| 产品名称 | 价格 | 指令 |
| DLD-1人结直肠腺癌上皮细胞(STR鉴定) | ¥1350.00 | 购物车 》 |
| NCI-H716 [H716]人结直肠腺癌细胞(STR鉴定) | ¥1800.00 | 购物车 》 |
| COLO 320DM人结直肠腺癌细胞(STR鉴定)[细胞+500ml专培套餐促销] | ¥980.00 | 购物车 》 |
| 支原体检测试剂盒(买二送一) | ¥800.00 | 购物车 》 |
| 国产转染试剂 | ¥800.00 | 购物车 》 |
| 慢病毒介导基因沉默或过表达 | ¥询价 | 购物车 》 |
| 绿/红色荧光蛋白标记 | ¥询价 | 购物车 》 |
| RPMI-1640基础培养基 | ¥56.00 | 购物车 》 |
| RPMI-1640完全培养基(10%FBS) | ¥350.00 | 购物车 》 |
 上海中乔新舟生物科技有限公司
上海中乔新舟生物科技有限公司