|
产品名称 |
RL95-2人子宫内膜癌细胞 |
|
货号 |
ZQ0362 |
|
产品介绍 |
RL95-2 是一种上皮样细胞,是从一名 65 岁白人癌症患者的子宫内膜中分离出来的。这些细胞有α角蛋白,定义明确的连接复合体,张力丝和表面微绒毛。
注意事项: |
|
种属 |
人 |
|
性别/年龄 |
女/65岁 |
|
组织 |
子宫;子宫内膜 |
|
疾病 |
癌 |
|
细胞类型 |
肿瘤细胞 |
|
形态学 |
上皮样/或圆形 |
|
生长方式 |
贴壁 |
|
倍增时间 |
大约22小时 |
|
培养基和添加剂 |
DMEM/F-12(中乔新舟 货号:ZQ-600)+10%胎牛血清(中乔新舟 货号:ZQ500-A)+1%P/S(中乔新舟 货号:CSP006)+0.005mg/mL胰岛素(中乔新舟 货号:CSP001-10) |
|
推荐完全培养基货号 |
|
|
生物安全等级 |
BSL-1 |
|
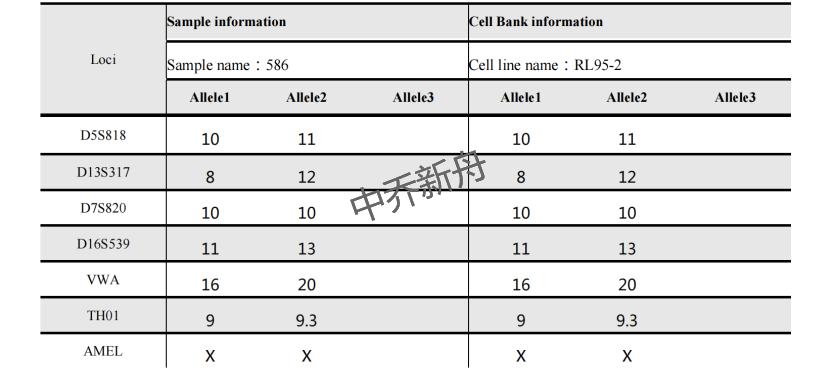
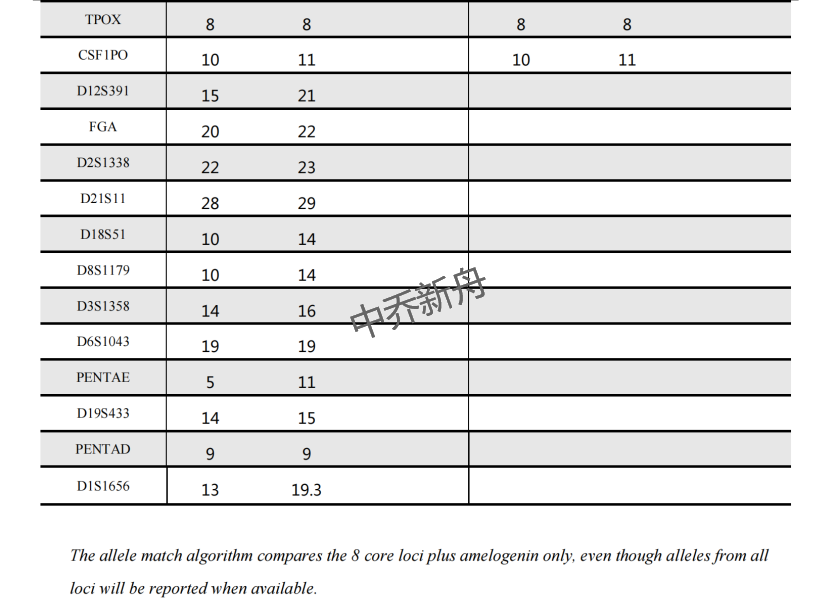
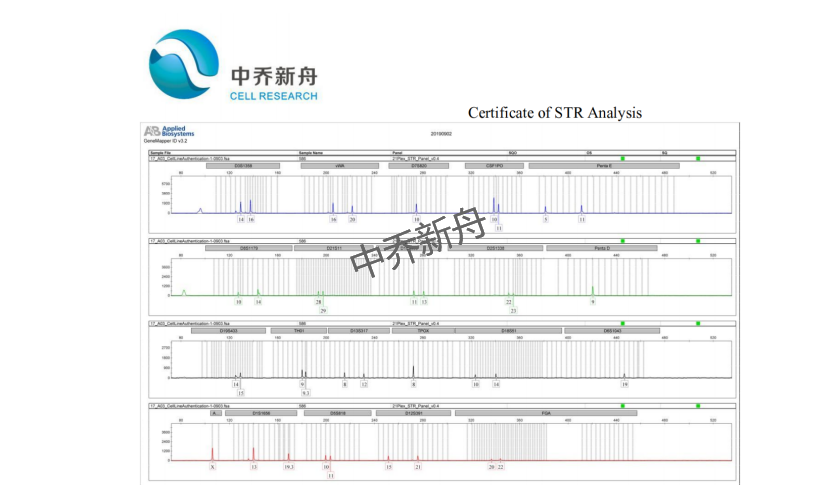
STR位点信息 |
Amelogenin: X CSF1PO: 10,11 D13S317: 8,12 D16S539: 11,13 D5S818: 10,11 D7S820: 10 TH01: 9,9.3 TPOX: 8 vWA: 16,20 |
|
培养条件 |
95%空气,5%二氧化碳;37℃ |
|
抗原表达/受体表达 |
*** |
|
基因表达 |
*** |
|
保藏机构 |
ATCC; CRL-1671;BCRJ 0366 |
|
供应限制 |
仅供科研使用 |
|
货号 |
ZQ0362 |
|
发货规格 |
活细胞:T25培养瓶*1瓶或者1ml 冻存管*2支(细胞量约为1x10^6 Cells/Vial)二选一 |
|
发货形式 |
活细胞:常温运输;冻存管:干冰运输 |
|
储存温度 |
活细胞:培养箱;冻存管:液氮罐 |
|
产地 |
中国 |
|
供应限制 |
仅供科研使用 |
论文标题: Dachsous cadherin related 1 (DCHS1) is a novel biomarker for immune infiltration and epithelial-mesenchymal transition in endometrial cancer via pan-cancer analysis
DOI:10.1186/s13048-024-01478-1.
发表时间: 2024-8-9
期刊: Journal of Ovarian Research
影响因子: 3.8
货号: ZQ0362
产品名称: RL95-2 cells
原文链接:https://ovarianresearch.biomedcentral.com/articles/10.1186/s13048-024-01478-1
论文标题: A flexible rapid self-assembly scaffold-net DNA hydrogel exhibiting cell mobility control
DOI: 10.1016/j.mtchem.2021.100680原文链接: https://www.sciencedirect.com/science/article/pii/S2468519421002603
论文标题: UBE2C is upregulated by estrogen and promotes epithelial-mesenchymal transition via p53 in endometrial cancer
DOI: 10.1158/1541-7786.MCR-19-0561
发表时间: 2019-01-01
期刊: MOLECULAR CANCER RESEARCH
影响因子: 4.484
货号: ZQ0362
产品名称: RL95-2 cells
原文链接: https://mcr.aacrjournals.org/content/18/2/204.abstract
PubMed=6339371; DOI=10.1007/BF02618053
Way D.L., Grosso D.S., Davis J.R., Surwit E.A., Christian C.D.
Characterization of a new human endometrial carcinoma (RL95-2) established in tissue culture.
In Vitro 19:147-158(1983)
PubMed=2436984; DOI=10.1016/0090-8258(87)90286-1
Noumoff J.S., Haydock S.W., Sachdeva R., Heyner S., Pritchard M.L.
Characteristics of cell lines derived from normal and malignant endometrial tissue.
Gynecol. Oncol. 27:141-149(1987)
PubMed=1541432; DOI=10.1016/0090-8258(92)90045-K
Rantanen V., Grenman S.E., Kulmala J., Salmi T., Grenman R.
Radiation sensitivity of endometrial carcinoma in vitro.
Gynecol. Oncol. 44:217-222(1992)
PubMed=7798295; DOI=10.1007/BF01194268
Rantanen V., Grenman S.E., Kulmala J., Alanen K., Lakkala T., Grenman R.
Sublethal damage repair after fractionated irradiation in endometrial cancer cell lines tested with the 96-well plate clonogenic assay.
J. Cancer Res. Clin. Oncol. 120:712-716(1994)
PubMed=8123477; DOI=10.1038/bjc.1994.87
Rantanen V., Grenman S.E., Kulmala J., Grenman R.
Comparative evaluation of cisplatin and carboplatin sensitivity in endometrial adenocarcinoma cell lines.
Br. J. Cancer 69:482-486(1994)
PubMed=9887230; DOI=10.1006/gyno.1998.5194
Rantanen V., Grenman S.E., Kurvinen K., Hietanen S., Raitanen M., Syrjanen S.M.
p53 mutations and presence of HPV DNA do not correlate with radiosensitivity of gynecological cancer cell lines.
Gynecol. Oncol. 71:352-358(1998)
PubMed=12893190; DOI=10.1016/S0090-8258(03)00335-4
Tanaka R., Saito T., Ashihara K., Nishimura M., Mizumoto H., Kudo R.
Three-dimensional coculture of endometrial cancer cells and fibroblasts in human placenta derived collagen sponges and expression matrix metalloproteinases in these cells.
Gynecol. Oncol. 90:297-304(2003)
PubMed=20164919; DOI=10.1038/nature08768
Bignell G.R., Greenman C.D., Davies H., Butler A.P., Edkins S., Andrews J.M., Buck G., Chen L., Beare D., Latimer C., Widaa S., Hinton J., Fahey C., Fu B.-Y., Swamy S., Dalgliesh G.L., Teh B.T., Deloukas P., Yang F.-T., Campbell P.J., Futreal P.A., Stratton M.R.
Signatures of mutation and selection in the cancer genome.
Nature 463:893-898(2010)
PubMed=20215515; DOI=10.1158/0008-5472.CAN-09-3458
Rothenberg S.M., Mohapatra G., Rivera M.N., Winokur D., Greninger P., Nitta M., Sadow P.M., Sooriyakumar G., Brannigan B.W., Ulman M.J., Perera R.M., Wang R., Tam A., Ma X.-J., Erlander M., Sgroi D.C., Rocco J.W., Lingen M.W., Cohen E.E.W., Louis D.N., Settleman J., Haber D.A.
A genome-wide screen for microdeletions reveals disruption of polarity complex genes in diverse human cancers.
Cancer Res. 70:2158-2164(2010)
PubMed=20944090; DOI=10.1126/scitranslmed.3001538
Dedes K.J., Wetterskog D., Mendes-Pereira A.M., Natrajan R., Lambros M.B., Geyer F.C., Vatcheva R., Savage K., Mackay A., Lord C.J., Ashworth A., Reis-Filho J.S.
PTEN deficiency in endometrioid endometrial adenocarcinomas predicts sensitivity to PARP inhibitors.
Sci. Transl. Med. 2:53ra75.1-53ra75.8(2010)
PubMed=22460905; DOI=10.1038/nature11003
Barretina J.G., Caponigro G., Stransky N., Venkatesan K., Margolin A.A., Kim S., Wilson C.J., Lehar J., Kryukov G.V., Sonkin D., Reddy A., Liu M., Murray L., Berger M.F., Monahan J.E., Morais P., Meltzer J., Korejwa A., Jane-Valbuena J., Mapa F.A., Thibault J., Bric-Furlong E., Raman P., Shipway A., Engels I.H., Cheng J., Yu G.-Y.K., Yu J.-J., Aspesi P. Jr., de Silva M., Jagtap K., Jones M.D., Wang L., Hatton C., Palescandolo E., Gupta S., Mahan S., Sougnez C., Onofrio R.C., Liefeld T., MacConaill L.E., Winckler W., Reich M., Li N.-X., Mesirov J.P., Gabriel S.B., Getz G., Ardlie K., Chan V., Myer V.E., Weber B.L., Porter J., Warmuth M., Finan P., Harris J.L., Meyerson M.L., Golub T.R., Morrissey M.P., Sellers W.R., Schlegel R., Garraway L.A.
The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity.
Nature 483:603-607(2012)
PubMed=22710073; DOI=10.1016/j.ygyno.2012.06.017
Korch C.T., Spillman M.A., Jackson T.A., Jacobsen B.M., Murphy S.K., Lessey B.A., Jordan V.C., Bradford A.P.
DNA profiling analysis of endometrial and ovarian cell lines reveals misidentification, redundancy and contamination.
Gynecol. Oncol. 127:241-248(2012)
PubMed=25877200; DOI=10.1038/nature14397
Yu M., Selvaraj S.K., Liang-Chu M.M.Y., Aghajani S., Busse M., Yuan J., Lee G., Peale F.V., Klijn C., Bourgon R., Kaminker J.S., Neve R.M.
A resource for cell line authentication, annotation and quality control.
Nature 520:307-311(2015)
PubMed=26589293; DOI=10.1186/s13073-015-0240-5
Scholtalbers J., Boegel S., Bukur T., Byl M., Goerges S., Sorn P., Loewer M., Sahin U., Castle J.C.
TCLP: an online cancer cell line catalogue integrating HLA type, predicted neo-epitopes, virus and gene expression.
Genome Med. 7:118.1-118.7(2015)
PubMed=27397505; DOI=10.1016/j.cell.2016.06.017
Iorio F., Knijnenburg T.A., Vis D.J., Bignell G.R., Menden M.P., Schubert M., Aben N., Goncalves E., Barthorpe S., Lightfoot H., Cokelaer T., Greninger P., van Dyk E., Chang H., de Silva H., Heyn H., Deng X.-M., Egan R.K., Liu Q.-S., Mironenko T., Mitropoulos X., Richardson L., Wang J.-H., Zhang T.-H., Moran S., Sayols S., Soleimani M., Tamborero D., Lopez-Bigas N., Ross-Macdonald P., Esteller M., Gray N.S., Haber D.A., Stratton M.R., Benes C.H., Wessels L.F.A., Saez-Rodriguez J., McDermott U., Garnett M.J.
A landscape of pharmacogenomic interactions in cancer.
Cell 166:740-754(2016)
PubMed=28196595; DOI=10.1016/j.ccell.2017.01.005
Li J., Zhao W., Akbani R., Liu W.-B., Ju Z.-L., Ling S.-Y., Vellano C.P., Roebuck P., Yu Q.-H., Eterovic A.K., Byers L.A., Davies M.A., Deng W.-L., Gopal Y.N.V., Chen G., von Euw E.M., Slamon D.J., Conklin D., Heymach J.V., Gazdar A.F., Minna J.D., Myers J.N., Lu Y.-L., Mills G.B., Liang H.
Characterization of human cancer cell lines by reverse-phase protein arrays.
Cancer Cell 31:225-239(2017)
PubMed=30894373; DOI=10.1158/0008-5472.CAN-18-2747
Dutil J., Chen Z.-H., Monteiro A.N.A., Teer J.K., Eschrich S.A.
An interactive resource to probe genetic diversity and estimated ancestry in cancer cell lines.
Cancer Res. 79:1263-1273(2019)
PubMed=30971826; DOI=10.1038/s41586-019-1103-9
Behan F.M., Iorio F., Picco G., Goncalves E., Beaver C.M., Migliardi G., Santos R., Rao Y., Sassi F., Pinnelli M., Ansari R., Harper S., Jackson D.A., McRae R., Pooley R., Wilkinson P., van der Meer D.J., Dow D., Buser-Doepner C.A., Bertotti A., Trusolino L., Stronach E.A., Saez-Rodriguez J., Yusa K., Garnett M.J.
Prioritization of cancer therapeutic targets using CRISPR-Cas9 screens.
Nature 568:511-516(2019)
PubMed=31068700; DOI=10.1038/s41586-019-1186-3
Ghandi M., Huang F.W., Jane-Valbuena J., Kryukov G.V., Lo C.C., McDonald E.R. III, Barretina J.G., Gelfand E.T., Bielski C.M., Li H.-X., Hu K., Andreev-Drakhlin A.Y., Kim J., Hess J.M., Haas B.J., Aguet F., Weir B.A., Rothberg M.V., Paolella B.R., Lawrence M.S., Akbani R., Lu Y.-L., Tiv H.L., Gokhale P.C., de Weck A., Mansour A.A., Oh C., Shih J., Hadi K., Rosen Y., Bistline J., Venkatesan K., Reddy A., Sonkin D., Liu M., Lehar J., Korn J.M., Porter D.A., Jones M.D., Golji J., Caponigro G., Taylor J.E., Dunning C.M., Creech A.L., Warren A.C., McFarland J.M., Zamanighomi M., Kauffmann A., Stransky N., Imielinski M., Maruvka Y.E., Cherniack A.D., Tsherniak A., Vazquez F., Jaffe J.D., Lane A.A., Weinstock D.M., Johannessen C.M., Morrissey M.P., Stegmeier F., Schlegel R., Hahn W.C., Getz G., Mills G.B., Boehm J.S., Golub T.R., Garraway L.A., Sellers W.R.
Next-generation characterization of the Cancer Cell Line Encyclopedia.
Nature 569:503-508(2019)
PubMed=31395879; DOI=10.1038/s41467-019-11415-2
Yu K., Chen B., Aran D., Charalel J., Yau C., Wolf D.M., van 't Veer L.J., Butte A.J., Goldstein T., Sirota M.
Comprehensive transcriptomic analysis of cell lines as models of primary tumors across 22 tumor types.
Nat. Commun. 10:3574.1-3574.11(2019)
PubMed=33174010; DOI=10.3892/ijo.2020.5139
Devor E.J., Gonzalez-Bosquet J., Thiel K.W., Leslie K.K.
Genomic characterization of five commonly used endometrial cancer cell lines.
Int. J. Oncol. 57:1348-1357(2020)
PubMed=35839778; DOI=10.1016/j.ccell.2022.06.010
Goncalves E., Poulos R.C., Cai Z.-X., Barthorpe S., Manda S.S., Lucas N., Beck A., Bucio-Noble D., Dausmann M., Hall C., Hecker M., Koh J., Lightfoot H., Mahboob S., Mali I., Morris J., Richardson L., Seneviratne A.J., Shepherd R., Sykes E., Thomas F., Valentini S., Williams S.G., Wu Y.-X., Xavier D., MacKenzie K.L., Hains P.G., Tully B., Robinson P.J., Zhong Q., Garnett M.J., Reddel R.R.
Pan-cancer proteomic map of 949 human cell lines.
Cancer Cell 40:835-849.e8(2022)
| 产品名称 | 价格 | 指令 |
| HEC-1-A人子宫内膜腺癌细胞(STR鉴定)[细胞+500ml专培套餐促销] | ¥980.00 | 购物车 》 |
| HEC-1-B人子宫内膜腺癌细胞(STR鉴定)[细胞+500ml专培套餐促销] | ¥980.00 | 购物车 》 |
| AN3CA人子宫内膜癌细胞 (STR鉴定)[细胞+500ml专培套餐促销] | ¥980 | 购物车 》 |
| 支原体检测试剂盒(买二送一) | ¥800.00 | 购物车 》 |
| 支原体清除试剂盒 | ¥600.00 | 购物车 》 |
| 国产转染试剂 | ¥800.00 | 购物车 》 |
| 国产优级胎牛血清 | ¥1800.00 | 购物车 》 |
| DMEM/F12基础培养基 | ¥56.00 | 购物车 》 |
| DMEM/F12完全培养基(10%FBS;Insulin) | ¥550.00 | 购物车 》 |
| 人源细胞STR鉴定服务 | ¥900.00 | 购物车 》 |
| ISK人子宫内膜癌细胞(STR鉴定)[细胞+500ml专培套餐促销] | ¥980 | 购物车 》 |
 上海中乔新舟生物科技有限公司
上海中乔新舟生物科技有限公司