|
产品名称 |
MDA-MB-415人乳腺腺癌细胞 |
|
货号 |
ZQ0354 |
|
产品介绍 |
这株细胞表达WNT7B癌基因。 8168088]. 带瘤患者来自巴拉圭,虽然填报的是白人,但细胞表型存在G6PD A型,显示其属于混血。细胞株形成平展延伸的上皮细胞样,在电镜下呈现结节,伴随着延伸的微管和微板。不容易用胰酶消化。
注意事项: |
|
种属 |
人 |
|
性别/年龄 |
女/38岁 |
|
组织 |
乳腺/乳房;来源于转移部位:胸腔积液 |
|
疾病 |
腺癌 |
|
细胞类型 |
肿瘤细胞 |
|
形态学 |
上皮的 |
|
生长方式 |
贴壁 |
|
倍增时间 |
大约88.87小时 |
|
培养基和添加剂(默认) |
DMEM (中乔新舟 货号:ZQ-100)+10%FBS(中乔新舟 货号:ZQ0500)+1%双抗(中乔新舟 货号:CSP006) 注意默认方案培养条件:95%空气,5%CO2;37℃ |
|
培养方案B(可选) |
L-15(中乔新舟 货号:ZQ-1100)+15%FBS(中乔新舟 货号:ZQ0500)+10ug/ml胰岛素(中乔新舟 货号:CSP001-10)+10ug/ml谷胱甘肽+1%P/S(中乔新舟 货号:CSP006) 注意B方案培养条件:100%空气;37℃ |
|
推荐默认完全培养基货号 |
|
|
B方案完全培养基货号 |
|
|
生物安全等级 |
BSL-1 |
|
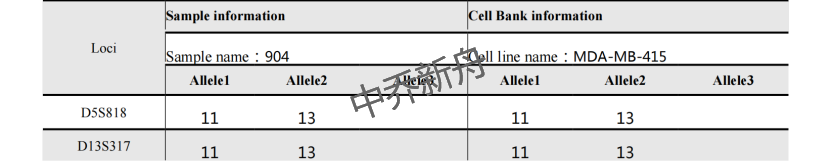
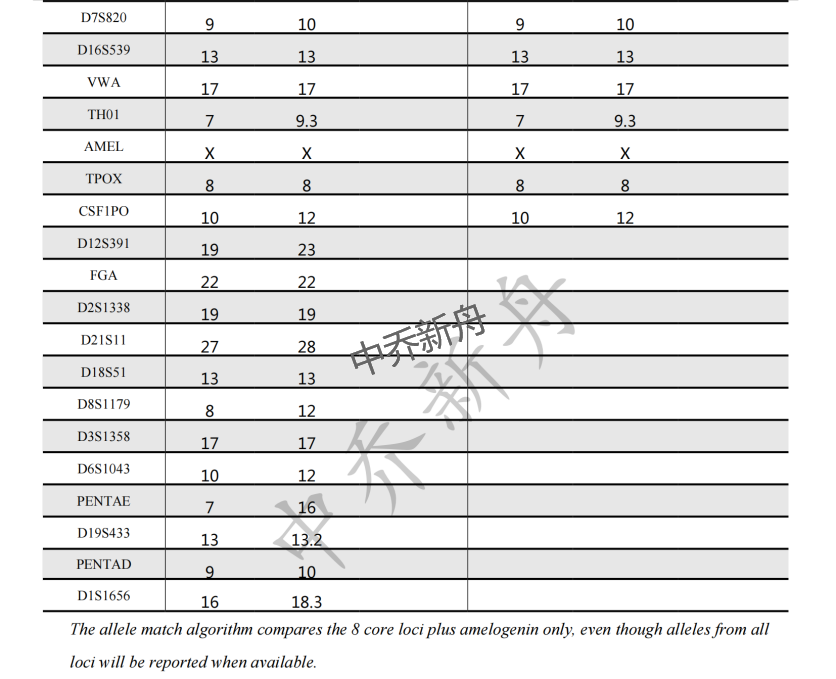
STR位点信息 |
Amelogenin: X CSF1PO: 10,12 D13S317: 11,13 D16S539: 13 D5S818: 11,13 D7S820: 9,10 TH01: 7,9.3 TPOX: 8 vWA: 17 |
|
默认方案培养条件 |
95%空气,5%CO2;37℃ |
|
抗原表达/受体表达 |
*** |
|
基因表达 |
*** |
|
保藏机构 |
ATCC; HTB-128 |
|
供应限制 |
仅供科研使用 |
|
货号 |
ZQ0354 |
|
发货规格 |
活细胞:T25培养瓶*1瓶或者1ml 冻存管*2支(细胞量约为1x10^6 Cells/Vial)二选一 |
|
发货形式 |
活细胞:常温运输;冻存管:干冰运输 |
|
储存温度 |
活细胞:培养箱;冻存管:液氮罐 |
|
产地 |
中国 |
|
供应限制 |
仅供科研使用 |
PubMed=730202; DOI=10.1007/BF02616120
Cailleau R.M., Olive M., Cruciger Q.V.J.
Long-term human breast carcinoma cell lines of metastatic origin: preliminary characterization.
In Vitro 14:911-915(1978)
PubMed=6935474; DOI=10.1093/jnci/66.2.239
Wright W.C., Daniels W.P., Fogh J.
Distinction of seventy-one cultured human tumor cell lines by polymorphic enzyme analysis.
J. Natl. Cancer Inst. 66:239-247(1981)
PubMed=7272986; DOI=10.1016/0165-4608(81)90057-1
Satya-Prakash K.L., Pathak S., Hsu T.-C., Olive M., Cailleau R.M.
Cytogenetic analysis on eight human breast tumor cell lines: high frequencies of 1q, 11q and HeLa-like marker chromosomes.
Cancer Genet. Cytogenet. 3:61-73(1981)
DOI=10.1016/B978-0-12-333530-2.50009-5
Leibovitz A.
Cell lines from human breast.
(In) Atlas of human tumor cell lines; Hay R.J., Park J.-G., Gazdar A.F. (eds.); pp.161-184; Academic Press; New York (1994)
PubMed=9331070
Teng D.H.-F., Perry W.L. III, Hogan J.K., Baumgard M.L., Bell R., Berry S., Davis T., Frank D., Frye C., Hattier T., Hu R., Jammulapati S., Janecki T., Leavitt A., Mitchell J.T., Pero R., Sexton D., Schroeder M., Su P.-H., Swedlund B., Kyriakis J.M., Avruch J., Bartel P., Wong A.K.C., Oliphant A., Thomas A., Skolnick M.H., Tavtigian S.V.
Human mitogen-activated protein kinase kinase 4 as a candidate tumor suppressor.
Cancer Res. 57:4177-4182(1997)
PubMed=10969801
Forozan F., Mahlamaki E.H., Monni O., Chen Y.-D., Veldman R., Jiang Y., Gooden G.C., Ethier S.P., Kallioniemi A., Kallioniemi O.-P.
Comparative genomic hybridization analysis of 38 breast cancer cell lines: a basis for interpreting complementary DNA microarray data.
Cancer Res. 60:4519-4525(2000)
PubMed=11343771; DOI=10.1016/S0165-4608(00)00387-3
Rummukainen J., Kytola S., Karhu R., Farnebo F., Larsson C., Isola J.J.
Aberrations of chromosome 8 in 16 breast cancer cell lines by comparative genomic hybridization, fluorescence in situ hybridization, and spectral karyotyping.
Cancer Genet. Cytogenet. 126:1-7(2001)
PubMed=12800145; DOI=10.1002/gcc.10218
Adelaide J., Huang H.-E., Murati A., Alsop A.E., Orsetti B., Mozziconacci M.-J., Popovici C., Ginestier C., Letessier A., Basset C., Courtay-Cahen C., Jacquemier J., Theillet C., Birnbaum D., Edwards P.A.W., Chaffanet M.
A recurrent chromosome translocation breakpoint in breast and pancreatic cancer cell lines targets the neuregulin/NRG1 gene.
Genes Chromosomes Cancer 37:333-345(2003)
PubMed=16397213; DOI=10.1158/0008-5472.CAN-05-2853
Elstrodt F., Hollestelle A., Nagel J.H.A., Gorin M., Wasielewski M., van den Ouweland A.M.W., Merajver S.D., Ethier S.P., Schutte M.
BRCA1 mutation analysis of 41 human breast cancer cell lines reveals three new deleterious mutants.
Cancer Res. 66:41-45(2006)
PubMed=16541312; DOI=10.1007/s10549-006-9186-z
Wasielewski M., Elstrodt F., Klijn J.G.M., Berns E.M.J.J., Schutte M.
Thirteen new p53 gene mutants identified among 41 human breast cancer cell lines.
Breast Cancer Res. Treat. 99:97-101(2006)
PubMed=17157791; DOI=10.1016/j.ccr.2006.10.008
Neve R.M., Chin K., Fridlyand J., Yeh J., Baehner F.L., Fevr T., Clark L., Bayani N., Coppe J.-P., Tong F., Speed T., Spellman P.T., DeVries S., Lapuk A., Wang N.J., Kuo W.-L., Stilwell J.L., Pinkel D., Albertson D.G., Waldman F.M., McCormick F., Dickson R.B., Johnson M.D., Lippman M.E., Ethier S.P., Gazdar A.F., Gray J.W.
A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes.
Cancer Cell 10:515-527(2006)
PubMed=18516279; DOI=10.1016/j.molonc.2007.02.004
Kenny P.A., Lee G.Y., Myers C.A., Neve R.M., Semeiks J.R., Spellman P.T., Lorenz K., Lee E.H., Barcellos-Hoff M.H., Petersen O.W., Gray J.W., Bissell M.J.
The morphologies of breast cancer cell lines in three-dimensional assays correlate with their profiles of gene expression.
Mol. Oncol. 1:84-96(2007)
PubMed=19593635; DOI=10.1007/s10549-009-0460-8
Hollestelle A., Nagel J.H.A., Smid M., Lam S., Elstrodt F., Wasielewski M., Ng S.S., French P.J., Peeters J.K., Rozendaal M.J., Riaz M., Koopman D.G., ten Hagen T.L.M., de Leeuw B.H.C.G.M., Zwarthoff E.C., Teunisse A., van der Spek P.J., Klijn J.G.M., Dinjens W.N.M., Ethier S.P., Clevers H.C., Jochemsen A.G., den Bakker M.A., Foekens J.A., Martens J.W.M., Schutte M.
Distinct gene mutation profiles among luminal-type and basal-type breast cancer cell lines.
Breast Cancer Res. Treat. 121:53-64(2010)
PubMed=20070913; DOI=10.1186/1471-2407-10-15
Tsuji K., Kawauchi S., Saito S., Furuya T., Ikemoto K., Nakao M., Yamamoto S., Oka M., Hirano T., Sasaki K.
Breast cancer cell lines carry cell line-specific genomic alterations that are distinct from aberrations in breast cancer tissues: comparison of the CGH profiles between cancer cell lines and primary cancer tissues.
BMC Cancer 10:15.1-15.10(2010)
PubMed=20164919; DOI=10.1038/nature08768
Bignell G.R., Greenman C.D., Davies H., Butler A.P., Edkins S., Andrews J.M., Buck G., Chen L., Beare D., Latimer C., Widaa S., Hinton J., Fahey C., Fu B.-Y., Swamy S., Dalgliesh G.L., Teh B.T., Deloukas P., Yang F.-T., Campbell P.J., Futreal P.A., Stratton M.R.
Signatures of mutation and selection in the cancer genome.
Nature 463:893-898(2010)
PubMed=22460905; DOI=10.1038/nature11003
Barretina J.G., Caponigro G., Stransky N., Venkatesan K., Margolin A.A., Kim S., Wilson C.J., Lehar J., Kryukov G.V., Sonkin D., Reddy A., Liu M., Murray L., Berger M.F., Monahan J.E., Morais P., Meltzer J., Korejwa A., Jane-Valbuena J., Mapa F.A., Thibault J., Bric-Furlong E., Raman P., Shipway A., Engels I.H., Cheng J., Yu G.-Y.K., Yu J.-J., Aspesi P. Jr., de Silva M., Jagtap K., Jones M.D., Wang L., Hatton C., Palescandolo E., Gupta S., Mahan S., Sougnez C., Onofrio R.C., Liefeld T., MacConaill L.E., Winckler W., Reich M., Li N.-X., Mesirov J.P., Gabriel S.B., Getz G., Ardlie K., Chan V., Myer V.E., Weber B.L., Porter J., Warmuth M., Finan P., Harris J.L., Meyerson M.L., Golub T.R., Morrissey M.P., Sellers W.R., Schlegel R., Garraway L.A.
The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity.
Nature 483:603-607(2012)
PubMed=23601657; DOI=10.1186/bcr3415
Riaz M., van Jaarsveld M.T.M., Hollestelle A., Prager-van der Smissen W.J.C., Heine A.A.J., Boersma A.W.M., Liu J.-J., Helmijr J.C.A., Ozturk B., Smid M., Wiemer E.A.C., Foekens J.A., Martens J.W.M.
miRNA expression profiling of 51 human breast cancer cell lines reveals subtype and driver mutation-specific miRNAs.
Breast Cancer Res. 15:R33.1-R33.17(2013)
PubMed=24094812; DOI=10.1016/j.ccr.2013.08.020
Timmerman L.A., Holton T., Yuneva M., Louie R.J., Padro M., Daemen A., Hu M., Chan D.A., Ethier S.P., van 't Veer L.J., Polyak K., McCormick F., Gray J.W.
Glutamine sensitivity analysis identifies the xCT antiporter as a common triple-negative breast tumor therapeutic target.
Cancer Cell 24:450-465(2013)
PubMed=24162158; DOI=10.1007/s10549-013-2743-3
Prat A., Karginova O., Parker J.S., Fan C., He X.-P., Bixby L.M., Harrell J.C., Roman E., Adamo B., Troester M.A., Perou C.M.
Characterization of cell lines derived from breast cancers and normal mammary tissues for the study of the intrinsic molecular subtypes.
Breast Cancer Res. Treat. 142:237-255(2013)
PubMed=24176112; DOI=10.1186/gb-2013-14-10-r110
Daemen A., Griffith O.L., Heiser L.M., Wang N.J., Enache O.M., Sanborn Z., Pepin F., Durinck S., Korkola J.E., Griffith M., Hur J.S., Huh N., Chung J., Cope L., Fackler M.J., Umbricht C.B., Sukumar S., Seth P., Sukhatme V.P., Jakkula L.R., Lu Y.-L., Mills G.B., Cho R.J., Collisson E.A., van 't Veer L.J., Spellman P.T., Gray J.W.
Modeling precision treatment of breast cancer.
Genome Biol. 14:R110.1-R110.14(2013)
PubMed=25485619; DOI=10.1038/nbt.3080
Klijn C., Durinck S., Stawiski E.W., Haverty P.M., Jiang Z.-S., Liu H.-B., Degenhardt J., Mayba O., Gnad F., Liu J.-F., Pau G., Reeder J., Cao Y., Mukhyala K., Selvaraj S.K., Yu M.-M., Zynda G.J., Brauer M.J., Wu T.D., Gentleman R.C., Manning G., Yauch R.L., Bourgon R., Stokoe D., Modrusan Z., Neve R.M., de Sauvage F.J., Settleman J., Seshagiri S., Zhang Z.-M.
A comprehensive transcriptional portrait of human cancer cell lines.
Nat. Biotechnol. 33:306-312(2015)
PubMed=25877200; DOI=10.1038/nature14397
Yu M., Selvaraj S.K., Liang-Chu M.M.Y., Aghajani S., Busse M., Yuan J., Lee G., Peale F.V., Klijn C., Bourgon R., Kaminker J.S., Neve R.M.
A resource for cell line authentication, annotation and quality control.
Nature 520:307-311(2015)
PubMed=26589293; DOI=10.1186/s13073-015-0240-5
Scholtalbers J., Boegel S., Bukur T., Byl M., Goerges S., Sorn P., Loewer M., Sahin U., Castle J.C.
TCLP: an online cancer cell line catalogue integrating HLA type, predicted neo-epitopes, virus and gene expression.
Genome Med. 7:118.1-118.7(2015)
PubMed=27397505; DOI=10.1016/j.cell.2016.06.017
Iorio F., Knijnenburg T.A., Vis D.J., Bignell G.R., Menden M.P., Schubert M., Aben N., Goncalves E., Barthorpe S., Lightfoot H., Cokelaer T., Greninger P., van Dyk E., Chang H., de Silva H., Heyn H., Deng X.-M., Egan R.K., Liu Q.-S., Mironenko T., Mitropoulos X., Richardson L., Wang J.-H., Zhang T.-H., Moran S., Sayols S., Soleimani M., Tamborero D., Lopez-Bigas N., Ross-Macdonald P., Esteller M., Gray N.S., Haber D.A., Stratton M.R., Benes C.H., Wessels L.F.A., Saez-Rodriguez J., McDermott U., Garnett M.J.
A landscape of pharmacogenomic interactions in cancer.
Cell 166:740-754(2016)
PubMed=28196595; DOI=10.1016/j.ccell.2017.01.005
Li J., Zhao W., Akbani R., Liu W.-B., Ju Z.-L., Ling S.-Y., Vellano C.P., Roebuck P., Yu Q.-H., Eterovic A.K., Byers L.A., Davies M.A., Deng W.-L., Gopal Y.N.V., Chen G., von Euw E.M., Slamon D.J., Conklin D., Heymach J.V., Gazdar A.F., Minna J.D., Myers J.N., Lu Y.-L., Mills G.B., Liang H.
Characterization of human cancer cell lines by reverse-phase protein arrays.
Cancer Cell 31:225-239(2017)
PubMed=28889351; DOI=10.1007/s10549-017-4496-x
Saunus J.M., Smart C.E., Kutasovic J.R., Johnston R.L., Kalita-de Croft P., Miranda M., Rozali E.N., Vargas A.C., Reid L.E., Lorsy E., Cocciardi S., Seidens T., McCart Reed A.E., Dalley A.J., Wockner L.F., Johnson J., Sarkar D., Askarian-Amiri M.E., Simpson P.T., Khanna K.K., Chenevix-Trench G., Al-Ejeh F., Lakhani S.R.
Multidimensional phenotyping of breast cancer cell lines to guide preclinical research.
Breast Cancer Res. Treat. 167:289-301(2018)
PubMed=30894373; DOI=10.1158/0008-5472.CAN-18-2747
Dutil J., Chen Z.-H., Monteiro A.N.A., Teer J.K., Eschrich S.A.
An interactive resource to probe genetic diversity and estimated ancestry in cancer cell lines.
Cancer Res. 79:1263-1273(2019)
PubMed=30971826; DOI=10.1038/s41586-019-1103-9
Behan F.M., Iorio F., Picco G., Goncalves E., Beaver C.M., Migliardi G., Santos R., Rao Y., Sassi F., Pinnelli M., Ansari R., Harper S., Jackson D.A., McRae R., Pooley R., Wilkinson P., van der Meer D.J., Dow D., Buser-Doepner C.A., Bertotti A., Trusolino L., Stronach E.A., Saez-Rodriguez J., Yusa K., Garnett M.J.
Prioritization of cancer therapeutic targets using CRISPR-Cas9 screens.
Nature 568:511-516(2019)
PubMed=31068700; DOI=10.1038/s41586-019-1186-3
Ghandi M., Huang F.W., Jane-Valbuena J., Kryukov G.V., Lo C.C., McDonald E.R. III, Barretina J.G., Gelfand E.T., Bielski C.M., Li H.-X., Hu K., Andreev-Drakhlin A.Y., Kim J., Hess J.M., Haas B.J., Aguet F., Weir B.A., Rothberg M.V., Paolella B.R., Lawrence M.S., Akbani R., Lu Y.-L., Tiv H.L., Gokhale P.C., de Weck A., Mansour A.A., Oh C., Shih J., Hadi K., Rosen Y., Bistline J., Venkatesan K., Reddy A., Sonkin D., Liu M., Lehar J., Korn J.M., Porter D.A., Jones M.D., Golji J., Caponigro G., Taylor J.E., Dunning C.M., Creech A.L., Warren A.C., McFarland J.M., Zamanighomi M., Kauffmann A., Stransky N., Imielinski M., Maruvka Y.E., Cherniack A.D., Tsherniak A., Vazquez F., Jaffe J.D., Lane A.A., Weinstock D.M., Johannessen C.M., Morrissey M.P., Stegmeier F., Schlegel R., Hahn W.C., Getz G., Mills G.B., Boehm J.S., Golub T.R., Garraway L.A., Sellers W.R.
Next-generation characterization of the Cancer Cell Line Encyclopedia.
Nature 569:503-508(2019)
PubMed=35839778; DOI=10.1016/j.ccell.2022.06.010
Goncalves E., Poulos R.C., Cai Z.-X., Barthorpe S., Manda S.S., Lucas N., Beck A., Bucio-Noble D., Dausmann M., Hall C., Hecker M., Koh J., Lightfoot H., Mahboob S., Mali I., Morris J., Richardson L., Seneviratne A.J., Shepherd R., Sykes E., Thomas F., Valentini S., Williams S.G., Wu Y.-X., Xavier D., MacKenzie K.L., Hains P.G., Tully B., Robinson P.J., Zhong Q., Garnett M.J., Reddel R.R.
Pan-cancer proteomic map of 949 human cell lines.
Cancer Cell 40:835-849.e8(2022)

| 产品名称 | 价格 | 指令 |
| MCF-7人乳腺癌细胞(STR鉴定)[细胞+500ml专培套餐促销] | ¥980 | 购物车 》 |
| MDA-MB-453人乳腺癌细胞(STR鉴定)[细胞+500ml专培套餐促销] | ¥980.00 | 购物车 》 |
| ZR-75-1人乳腺癌细胞(STR鉴定) | ¥1600.00 | 购物车 》 |
| SHZ-88大鼠乳腺癌细胞(种属鉴定)[细胞+500ml专培套餐促销] | ¥980.00 | 购物车 》 |
| 人乳腺癌细胞BcaP-37(hela污染) | ¥1200.00 | 购物车 》 |
| MDA-MB-231人乳腺癌细胞(STR鉴定)[细胞+500ml专培套餐促销] | ¥980 | 购物车 》 |
| 4T1小鼠乳腺癌细胞(种属鉴定)[细胞+500ml专培套餐促销] | ¥980 | 购物车 》 |
| ZR-75-30人乳腺癌细胞(STR鉴定) | ¥1800.00 | 购物车 》 |
| HCC1937人乳腺癌细胞(STR鉴定)[细胞+500ml专培套餐促销] | ¥980.00 | 购物车 》 |
| Hs 578T人乳腺癌细胞(STR鉴定)[细胞+500ml专培套餐促销] | ¥980.00 | 购物车 》 |
| MDA-MB-468人乳腺癌细胞(STR鉴定)[细胞+500ml专培套餐促销] | ¥980 | 购物车 》 |
| 支原体检测试剂盒(买二送一) | ¥800.00 | 购物车 》 |
| 支原体清除试剂盒 | ¥600.00 | 购物车 》 |
| 国产转染试剂 | ¥800.00 | 购物车 》 |
| 优级胎牛血清 | ¥2580(开学促销) | 购物车 》 |
| L-15 基础培养基 | ¥158.00 | 购物车 》 |
| L-15完全培养基(15%FBS;10ug/ml Insulin;10ug/ml glutathione) | ¥980.00 | 购物车 》 |
 上海中乔新舟生物科技有限公司
上海中乔新舟生物科技有限公司